Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2885-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2885-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 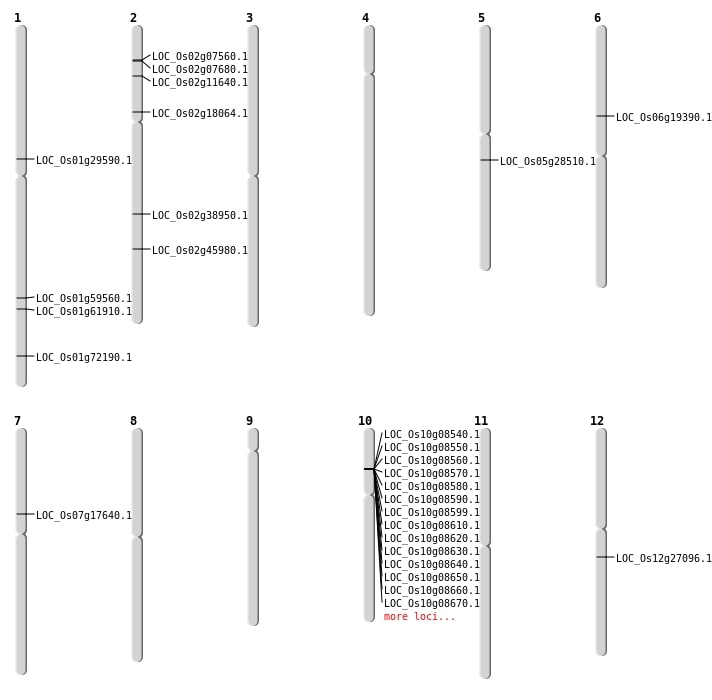
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13018313 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 16580755 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g29590 |
| SBS | Chr1 | 6988151 | A-C | HET | INTRON | |
| SBS | Chr10 | 14414328 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 15739419 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 21563933 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 22235822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 4819498 | G-A | HET | INTRON | |
| SBS | Chr11 | 5165496 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr11 | 5705102 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 21615320 | G-A | HET | INTRON | |
| SBS | Chr12 | 9728440 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 20090426 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 23539187 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g38950 |
| SBS | Chr2 | 2422831 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 5979100 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g11640 |
| SBS | Chr2 | 5979103 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 12550076 | T-A | HOMO | INTRON | |
| SBS | Chr3 | 1464969 | G-A | HET | INTRON | |
| SBS | Chr3 | 31435133 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 5180826 | A-C | HET | UTR_3_PRIME | |
| SBS | Chr4 | 1218218 | T-G | HET | INTRON | |
| SBS | Chr4 | 29736574 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 21572118 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 7555367 | T-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 8233504 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 14329984 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 19314314 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 6994945 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 10420379 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17640 |
| SBS | Chr7 | 11878498 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 456654 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 4996468 | T-A | HOMO | INTRON | |
| SBS | Chr8 | 21880396 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 24435829 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 24602772 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 25606796 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 1413980 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 20213487 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 20728514 | G-A | HET | INTERGENIC |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 112525 | 112539 | 14 | |
| Deletion | Chr4 | 156090 | 156097 | 7 | |
| Deletion | Chr5 | 2207251 | 2207256 | 5 | |
| Deletion | Chr2 | 3913400 | 3913402 | 2 | LOC_Os02g07560 |
| Deletion | Chr2 | 3996203 | 3996204 | 1 | LOC_Os02g07680 |
| Deletion | Chr10 | 4622001 | 4743000 | 120999 | 22 |
| Deletion | Chr10 | 6007258 | 6007264 | 6 | |
| Deletion | Chr4 | 6670807 | 6670808 | 1 | |
| Deletion | Chr5 | 7650592 | 7650598 | 6 | |
| Deletion | Chr11 | 9999908 | 9999921 | 13 | |
| Deletion | Chr3 | 10781447 | 10781448 | 1 | |
| Deletion | Chr2 | 11030303 | 11030304 | 1 | |
| Deletion | Chr6 | 11041676 | 11041683 | 7 | LOC_Os06g19390 |
| Deletion | Chr7 | 11824043 | 11824044 | 1 | |
| Deletion | Chr4 | 12486009 | 12486011 | 2 | |
| Deletion | Chr6 | 12550132 | 12550150 | 18 | |
| Deletion | Chr1 | 13404952 | 13404955 | 3 | |
| Deletion | Chr5 | 13503610 | 13503615 | 5 | |
| Deletion | Chr1 | 15591771 | 15591773 | 2 | |
| Deletion | Chr12 | 15881172 | 15881180 | 8 | LOC_Os12g27096 |
| Deletion | Chr5 | 16699410 | 16699414 | 4 | LOC_Os05g28510 |
| Deletion | Chr3 | 19681642 | 19681645 | 3 | |
| Deletion | Chr11 | 20132294 | 20132311 | 17 | |
| Deletion | Chr11 | 24852274 | 24852287 | 13 | |
| Deletion | Chr4 | 25979588 | 25979592 | 4 | |
| Deletion | Chr6 | 27463952 | 27463959 | 7 | |
| Deletion | Chr1 | 27661708 | 27661716 | 8 | |
| Deletion | Chr2 | 28018398 | 28018405 | 7 | LOC_Os02g45980 |
| Deletion | Chr4 | 28593946 | 28593953 | 7 | |
| Deletion | Chr3 | 28683435 | 28683437 | 2 | |
| Deletion | Chr4 | 32622327 | 32622331 | 4 | |
| Deletion | Chr3 | 33157737 | 33157741 | 4 | |
| Deletion | Chr4 | 33957516 | 33957526 | 10 | |
| Deletion | Chr1 | 34440210 | 34440222 | 12 | LOC_Os01g59560 |
| Deletion | Chr1 | 35835010 | 35835017 | 7 | LOC_Os01g61910 |
| Deletion | Chr1 | 36954603 | 36954604 | 1 | |
| Deletion | Chr1 | 42025462 | 42025487 | 25 |
Insertions: 8
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2032376 | Chr1 | 41863432 | LOC_Os01g72190 |
| Translocation | Chr4 | 7871808 | Chr2 | 34491837 | |
| Translocation | Chr12 | 12555239 | Chr1 | 1692190 | |
| Translocation | Chr12 | 15238163 | Chr2 | 10483612 | LOC_Os02g18064 |
| Translocation | Chr12 | 15244369 | Chr2 | 10483434 | LOC_Os02g18064 |
