Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2891-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2891-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 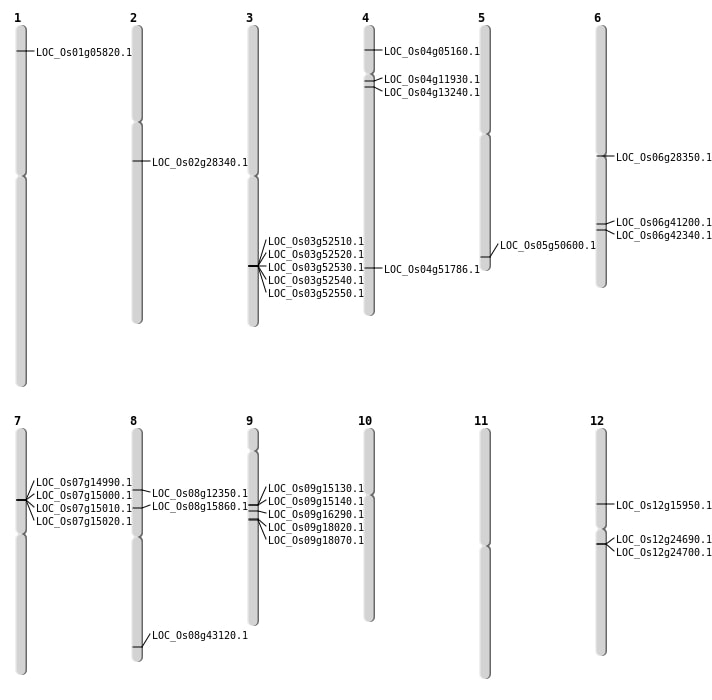
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22313842 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29099985 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29210117 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 5092598 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 13352660 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 4462553 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 12117870 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 2057264 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 21766548 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 3291165 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 23369682 | G-A | HET | INTRON | |
| SBS | Chr2 | 32319213 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 34820028 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 4228468 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 20462487 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 1304862 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 13440818 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 6735035 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 8107248 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 13122448 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 23534059 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 29004962 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g50600 |
| SBS | Chr5 | 7663366 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 11202939 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 19623274 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 6964862 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 7279811 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12350 |
| SBS | Chr9 | 1238183 | A-G | HOMO | INTRON |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 2502188 | 2502190 | 2 | |
| Deletion | Chr4 | 2571072 | 2571073 | 1 | LOC_Os04g05160 |
| Deletion | Chr7 | 2764256 | 2764258 | 2 | |
| Deletion | Chr4 | 2863397 | 2863412 | 15 | |
| Deletion | Chr3 | 2894085 | 2894086 | 1 | |
| Deletion | Chr3 | 5194385 | 5194399 | 14 | |
| Deletion | Chr4 | 6530001 | 6548000 | 17999 | LOC_Os04g11930 |
| Deletion | Chr4 | 7333672 | 7333680 | 8 | LOC_Os04g13240 |
| Deletion | Chr12 | 8395674 | 8395680 | 6 | |
| Deletion | Chr6 | 8460545 | 8460546 | 1 | |
| Deletion | Chr7 | 8593001 | 8614000 | 20999 | 4 |
| Deletion | Chr12 | 9089519 | 9089526 | 7 | LOC_Os12g15950 |
| Deletion | Chr9 | 9170001 | 9190000 | 19999 | 3 |
| Deletion | Chr7 | 9235565 | 9235570 | 5 | |
| Deletion | Chr11 | 9511585 | 9511589 | 4 | |
| Deletion | Chr8 | 9576001 | 9594000 | 17999 | LOC_Os08g15860 |
| Deletion | Chr5 | 9896915 | 9896916 | 1 | |
| Deletion | Chr6 | 10246262 | 10246263 | 1 | |
| Deletion | Chr9 | 10345091 | 10345094 | 3 | |
| Deletion | Chr5 | 10874574 | 10874575 | 1 | |
| Deletion | Chr9 | 11027001 | 11051000 | 23999 | 2 |
| Deletion | Chr12 | 12919640 | 12919643 | 3 | |
| Deletion | Chr1 | 13967298 | 13967299 | 1 | |
| Deletion | Chr12 | 14140001 | 14162000 | 21999 | 2 |
| Deletion | Chr5 | 15928692 | 15928711 | 19 | |
| Deletion | Chr6 | 16127347 | 16127348 | 1 | LOC_Os06g28350 |
| Deletion | Chr2 | 16750475 | 16750481 | 6 | LOC_Os02g28340 |
| Deletion | Chr1 | 17004384 | 17004394 | 10 | |
| Deletion | Chr8 | 18992263 | 18992264 | 1 | |
| Deletion | Chr10 | 21941471 | 21941473 | 2 | |
| Deletion | Chr2 | 22307213 | 22307220 | 7 | |
| Deletion | Chr6 | 24646001 | 24661000 | 14999 | 2 |
| Deletion | Chr5 | 25414028 | 25414029 | 1 | |
| Deletion | Chr1 | 25823676 | 25823677 | 1 | |
| Deletion | Chr8 | 26466001 | 26481000 | 14999 | LOC_Os08g43120 |
| Deletion | Chr6 | 26540266 | 26540274 | 8 | |
| Deletion | Chr7 | 28025231 | 28025233 | 2 | |
| Deletion | Chr3 | 30128001 | 30148000 | 19999 | 5 |
Insertions: 21
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 20117306 | 20547692 | |
| Inversion | Chr12 | 21731437 | 21784966 | |
| Inversion | Chr12 | 21731446 | 21784978 | |
| Inversion | Chr11 | 22238932 | 22649100 | |
| Inversion | Chr11 | 22239280 | 22686831 | |
| Inversion | Chr2 | 29037884 | 29831980 | |
| Inversion | Chr4 | 30686209 | 30770987 | LOC_Os04g51786 |
| Inversion | Chr4 | 30686220 | 30770996 | LOC_Os04g51786 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 3105428 | Chr3 | 25259158 | |
| Translocation | Chr3 | 24819508 | Chr1 | 2790235 | LOC_Os01g05820 |
