Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2895-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2895-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 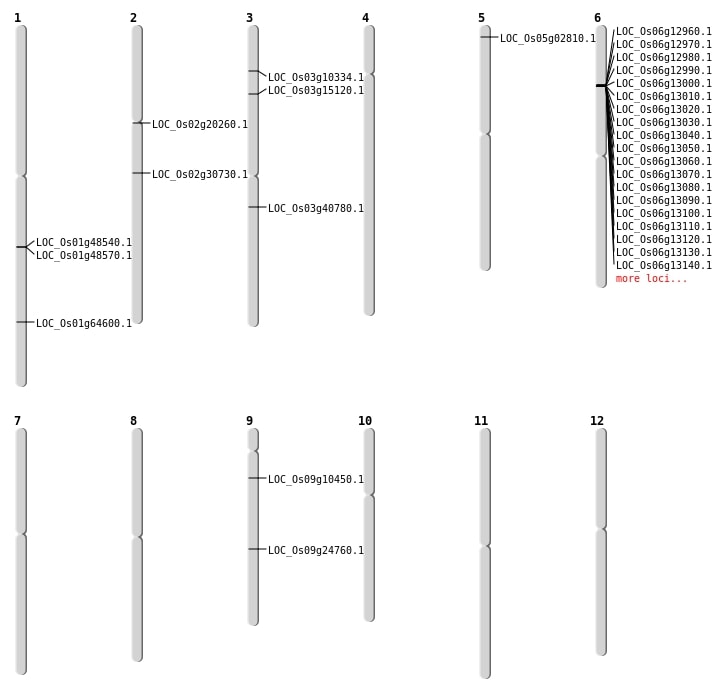
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr11 | 1483523 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 23779575 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 35023462 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 4183787 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 22671107 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g40780 |
| SBS | Chr3 | 23783851 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 5271418 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g10334 |
| SBS | Chr3 | 8260727 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15120 |
| SBS | Chr4 | 25226347 | G-A | HET | INTRON | |
| SBS | Chr4 | 32781000 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 34109512 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 20647444 | T-G | HET | INTRON | |
| SBS | Chr6 | 11334885 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11334886 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 28201553 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 4052973 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 4667803 | C-G | HET | INTRON | |
| SBS | Chr7 | 12378092 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 1586084 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17511197 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 17610008 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 18752262 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21086638 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 7031246 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 21750557 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 14752688 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g24760 |
| SBS | Chr9 | 5711829 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10450 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1788947 | 1788948 | 1 | |
| Deletion | Chr10 | 2722884 | 2722909 | 25 | |
| Deletion | Chr10 | 4808960 | 4808970 | 10 | |
| Deletion | Chr6 | 7088001 | 7285000 | 196999 | 31 |
| Deletion | Chr6 | 7894842 | 7894866 | 24 | LOC_Os06g14150 |
| Deletion | Chr2 | 9439103 | 9439105 | 2 | |
| Deletion | Chr2 | 11930204 | 11930205 | 1 | LOC_Os02g20260 |
| Deletion | Chr2 | 12455477 | 12455480 | 3 | |
| Deletion | Chr2 | 13787625 | 13787626 | 1 | |
| Deletion | Chr12 | 18996140 | 18996141 | 1 | |
| Deletion | Chr10 | 19821945 | 19821946 | 1 | |
| Deletion | Chr9 | 21429585 | 21429586 | 1 | |
| Deletion | Chr6 | 21967735 | 21967759 | 24 | |
| Deletion | Chr12 | 25326355 | 25326362 | 7 | |
| Deletion | Chr5 | 26013397 | 26013398 | 1 | |
| Deletion | Chr5 | 28439124 | 28439131 | 7 | |
| Deletion | Chr1 | 34270916 | 34270917 | 1 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 13763923 | 13763923 | 1 | |
| Insertion | Chr1 | 2133681 | 2133681 | 1 | |
| Insertion | Chr1 | 37492112 | 37492115 | 4 | LOC_Os01g64600 |
| Insertion | Chr10 | 7080943 | 7080943 | 1 | |
| Insertion | Chr11 | 16006926 | 16006927 | 2 | |
| Insertion | Chr11 | 16714835 | 16714836 | 2 | |
| Insertion | Chr12 | 13935481 | 13935481 | 1 | |
| Insertion | Chr12 | 15920631 | 15920631 | 1 | |
| Insertion | Chr2 | 11922011 | 11922013 | 3 | |
| Insertion | Chr2 | 9609950 | 9609965 | 16 | |
| Insertion | Chr5 | 11103006 | 11103016 | 11 | |
| Insertion | Chr6 | 30585089 | 30585089 | 1 | |
| Insertion | Chr8 | 22800889 | 22800889 | 1 | |
| Insertion | Chr8 | 2754604 | 2754604 | 1 | |
| Insertion | Chr9 | 111531 | 111532 | 2 | |
| Insertion | Chr9 | 21429693 | 21429693 | 1 | |
| Insertion | Chr9 | 21573886 | 21574005 | 120 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 147509 | 272435 | |
| Inversion | Chr5 | 1010885 | 1684505 | LOC_Os05g02810 |
| Inversion | Chr5 | 1398841 | 1684504 | |
| Inversion | Chr12 | 15182647 | 15321947 | |
| Inversion | Chr7 | 23070367 | 23244728 | |
| Inversion | Chr7 | 23070386 | 23244780 | |
| Inversion | Chr6 | 23720891 | 23930688 | LOC_Os06g40190 |
| Inversion | Chr1 | 42325652 | 42624478 | LOC_Os01g73570 |
| Inversion | Chr1 | 42325657 | 42624490 | LOC_Os01g73570 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 44561 | Chr4 | 7742301 | |
| Translocation | Chr9 | 44594 | Chr4 | 7547059 | |
| Translocation | Chr11 | 147514 | Chr1 | 27841319 | LOC_Os01g48540 |
| Translocation | Chr11 | 272437 | Chr1 | 27852507 | LOC_Os01g48570 |
| Translocation | Chr7 | 1583570 | Chr6 | 23720914 | |
| Translocation | Chr7 | 1589713 | Chr6 | 23930701 | LOC_Os06g40190 |
| Translocation | Chr7 | 1969104 | Chr2 | 20513513 | |
| Translocation | Chr11 | 6602818 | Chr2 | 4170016 | |
| Translocation | Chr11 | 26738186 | Chr2 | 18311741 | LOC_Os02g30730 |
| Translocation | Chr8 | 27374378 | Chr6 | 2829531 |
