Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2897-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2897-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 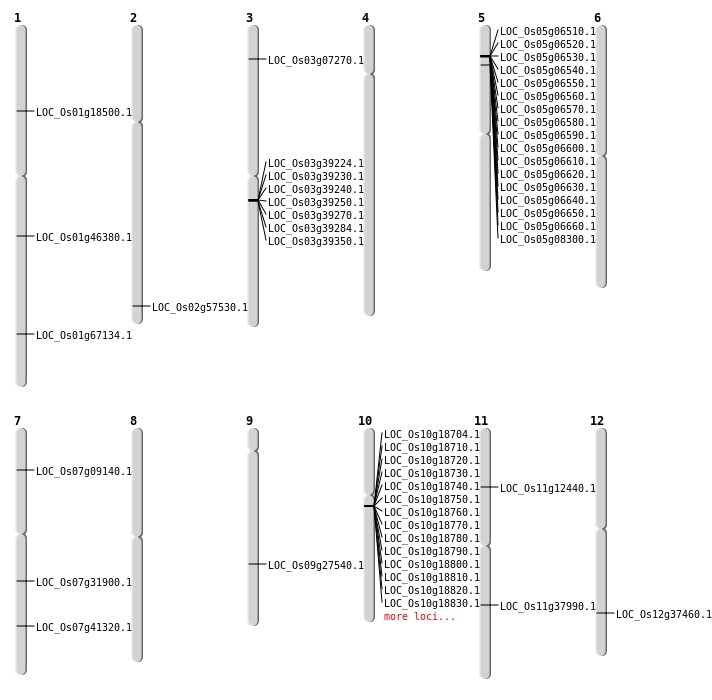
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10404460 | A-G | HET | SPLICE_SITE_DONOR | LOC_Os01g18500 |
| SBS | Chr1 | 26394294 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g46380 |
| SBS | Chr1 | 28962461 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 11348269 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g21980 |
| SBS | Chr10 | 11348270 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g21980 |
| SBS | Chr10 | 12914598 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 7854407 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15196680 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 17827027 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 24648405 | A-T | HET | INTRON | |
| SBS | Chr12 | 11877891 | C-T | HET | INTRON | |
| SBS | Chr12 | 15260540 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 10970346 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 21249331 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 4063767 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 14547660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 1998944 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 27644425 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 34452790 | A-T | HET | INTRON | |
| SBS | Chr5 | 26223390 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 26668448 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 3579783 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 22841878 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 5481990 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 21715954 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 7188318 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7758335 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 8015051 | C-T | HET | INTRON | |
| SBS | Chr8 | 15307845 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 6762455 | G-T | HET | INTRON | |
| SBS | Chr8 | 6762456 | G-T | HET | INTRON | |
| SBS | Chr9 | 16723758 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 3987949 | C-T | HET | INTRON |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1959100 | 1959102 | 2 | |
| Deletion | Chr5 | 3345001 | 3457000 | 111999 | 17 |
| Deletion | Chr5 | 3571557 | 3571560 | 3 | |
| Deletion | Chr3 | 3707691 | 3707751 | 60 | LOC_Os03g07270 |
| Deletion | Chr5 | 3936927 | 3936929 | 2 | |
| Deletion | Chr8 | 5915829 | 5915832 | 3 | |
| Deletion | Chr8 | 6240565 | 6240584 | 19 | |
| Deletion | Chr10 | 6695189 | 6695273 | 84 | |
| Deletion | Chr11 | 6844094 | 6844105 | 11 | |
| Deletion | Chr11 | 6950150 | 6950153 | 3 | LOC_Os11g12440 |
| Deletion | Chr9 | 6953482 | 6953485 | 3 | |
| Deletion | Chr3 | 7300112 | 7300113 | 1 | |
| Deletion | Chr2 | 8468172 | 8468176 | 4 | |
| Deletion | Chr4 | 9061678 | 9061679 | 1 | |
| Deletion | Chr10 | 9503001 | 9601000 | 97999 | 15 |
| Deletion | Chr8 | 11002611 | 11002629 | 18 | |
| Deletion | Chr5 | 11598476 | 11598479 | 3 | |
| Deletion | Chr4 | 12787751 | 12787752 | 1 | |
| Deletion | Chr5 | 12901257 | 12901264 | 7 | |
| Deletion | Chr8 | 13792219 | 13792279 | 60 | |
| Deletion | Chr1 | 16404901 | 16404910 | 9 | |
| Deletion | Chr10 | 16754143 | 16754154 | 11 | |
| Deletion | Chr4 | 18726926 | 18726938 | 12 | |
| Deletion | Chr4 | 18884108 | 18884112 | 4 | |
| Deletion | Chr7 | 18968654 | 18968710 | 56 | LOC_Os07g31900 |
| Deletion | Chr5 | 19696785 | 19696786 | 1 | |
| Deletion | Chr2 | 20287278 | 20287282 | 4 | |
| Deletion | Chr1 | 20899033 | 20899061 | 28 | |
| Deletion | Chr3 | 21780001 | 21825000 | 44999 | 7 |
| Deletion | Chr11 | 21849129 | 21849130 | 1 | |
| Deletion | Chr1 | 22572221 | 22572222 | 1 | |
| Deletion | Chr7 | 24756502 | 24756503 | 1 | LOC_Os07g41320 |
| Deletion | Chr4 | 26050638 | 26050639 | 1 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr5 | 29849191 | 29849899 | 708 | |
| Deletion | Chr2 | 35253305 | 35253311 | 6 | LOC_Os02g57530 |
| Deletion | Chr1 | 37209665 | 37209686 | 21 | |
| Deletion | Chr1 | 37209759 | 37209762 | 3 |
Insertions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10348131 | 10348133 | 3 | |
| Insertion | Chr10 | 11844458 | 11844458 | 1 | |
| Insertion | Chr10 | 14002198 | 14002198 | 1 | |
| Insertion | Chr10 | 3244889 | 3244889 | 1 | |
| Insertion | Chr10 | 6722414 | 6722437 | 24 | |
| Insertion | Chr10 | 9023677 | 9023680 | 4 | |
| Insertion | Chr11 | 14583272 | 14583294 | 23 | |
| Insertion | Chr11 | 5292125 | 5292126 | 2 | |
| Insertion | Chr12 | 16798949 | 16798952 | 4 | |
| Insertion | Chr12 | 22991616 | 22991620 | 5 | LOC_Os12g37460 |
| Insertion | Chr12 | 2998730 | 2998730 | 1 | |
| Insertion | Chr2 | 4467456 | 4467511 | 56 | |
| Insertion | Chr3 | 27940344 | 27940344 | 1 | |
| Insertion | Chr3 | 28982499 | 28982500 | 2 | |
| Insertion | Chr4 | 28820146 | 28820146 | 1 | |
| Insertion | Chr4 | 31890704 | 31890704 | 1 | |
| Insertion | Chr5 | 11574713 | 11574713 | 1 | |
| Insertion | Chr5 | 20416205 | 20416205 | 1 | |
| Insertion | Chr6 | 997606 | 997696 | 91 | |
| Insertion | Chr7 | 11302752 | 11302752 | 1 | |
| Insertion | Chr8 | 12820254 | 12820254 | 1 | |
| Insertion | Chr8 | 20767372 | 20767372 | 1 | |
| Insertion | Chr8 | 25123692 | 25123742 | 51 | |
| Insertion | Chr8 | 5291837 | 5291837 | 1 | |
| Insertion | Chr9 | 14772197 | 14772197 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 9502441 | 9678858 | |
| Inversion | Chr10 | 9597003 | 9679168 | |
| Inversion | Chr9 | 16725793 | 16733745 | LOC_Os09g27540 |
| Inversion | Chr9 | 16725797 | 16733753 | LOC_Os09g27540 |
| Inversion | Chr3 | 21241044 | 21820094 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1076403 | Chr1 | 30658065 | |
| Translocation | Chr6 | 3024365 | Chr2 | 16275481 | |
| Translocation | Chr7 | 4773878 | Chr4 | 4289191 | |
| Translocation | Chr7 | 4774280 | Chr4 | 4289185 | LOC_Os07g09140 |
| Translocation | Chr10 | 5409188 | Chr4 | 2632481 | |
| Translocation | Chr8 | 5495784 | Chr2 | 10021553 | |
| Translocation | Chr10 | 9678868 | Chr1 | 38974461 | LOC_Os01g67134 |
| Translocation | Chr10 | 9679152 | Chr1 | 38974461 | LOC_Os01g67134 |
| Translocation | Chr4 | 19009544 | Chr2 | 17021801 | |
| Translocation | Chr8 | 19389187 | Chr4 | 18052947 | |
| Translocation | Chr11 | 22529379 | Chr9 | 902063 | LOC_Os11g37990 |
