Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2914-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2914-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 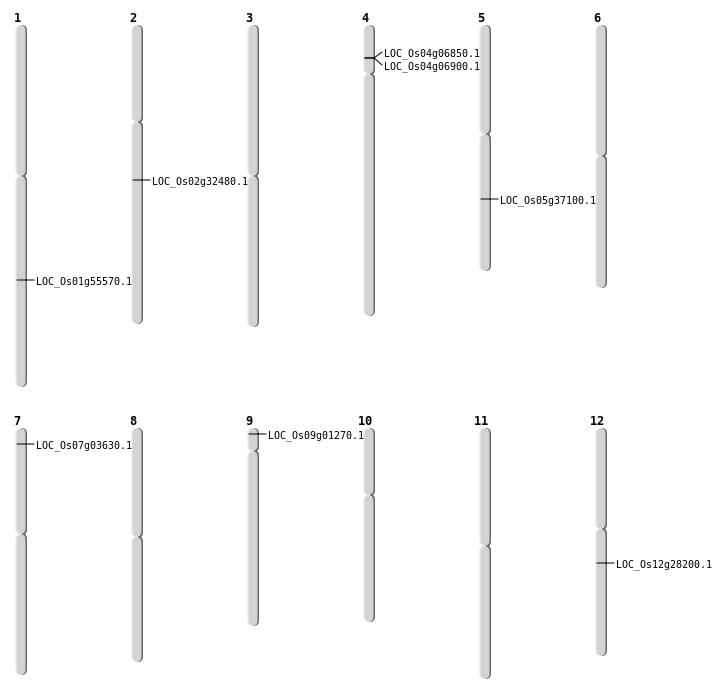
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18464457 | A-G | HOMO | INTRON | |
| SBS | Chr1 | 22563805 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 12646516 | T-C | HET | INTRON | |
| SBS | Chr10 | 19218738 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr11 | 12616298 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 14306669 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 26112826 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 8554777 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 16652317 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g28200 |
| SBS | Chr12 | 21092116 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 12245173 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 33462904 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 8102631 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 13151006 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 13559330 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 18488332 | G-C | HET | UTR_5_PRIME | |
| SBS | Chr3 | 18488333 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 22172838 | T-G | HET | UTR_5_PRIME | |
| SBS | Chr3 | 34367057 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 34467223 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 10578278 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 16853862 | G-A | HET | INTRON | |
| SBS | Chr4 | 21054826 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 28831824 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 3002231 | G-C | HOMO | INTRON | |
| SBS | Chr4 | 8934072 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 11748111 | G-A | HET | INTRON | |
| SBS | Chr6 | 17425811 | T-G | HET | INTRON | |
| SBS | Chr7 | 11178152 | G-A | HET | INTRON | |
| SBS | Chr7 | 8541666 | C-G | HET | INTRON | |
| SBS | Chr8 | 17564703 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 20468309 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 22635926 | C-T | HOMO | START_GAINED |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 234516 | 234576 | 60 | LOC_Os09g01270 |
| Deletion | Chr10 | 528885 | 528892 | 7 | |
| Deletion | Chr7 | 1463122 | 1463123 | 1 | LOC_Os07g03630 |
| Deletion | Chr2 | 1617243 | 1617245 | 2 | |
| Deletion | Chr1 | 2238991 | 2238999 | 8 | |
| Deletion | Chr5 | 2949906 | 2949907 | 1 | |
| Deletion | Chr4 | 3636525 | 3636526 | 1 | LOC_Os04g06900 |
| Deletion | Chr11 | 4886885 | 4886886 | 1 | |
| Deletion | Chr8 | 6084507 | 6084508 | 1 | |
| Deletion | Chr2 | 7522138 | 7522140 | 2 | |
| Deletion | Chr6 | 7638013 | 7638018 | 5 | |
| Deletion | Chr7 | 7819269 | 7819270 | 1 | |
| Deletion | Chr9 | 8926244 | 8926245 | 1 | |
| Deletion | Chr3 | 9118757 | 9118759 | 2 | |
| Deletion | Chr9 | 9165435 | 9165525 | 90 | |
| Deletion | Chr1 | 12715900 | 12715901 | 1 | |
| Deletion | Chr3 | 13150959 | 13150962 | 3 | |
| Deletion | Chr3 | 14313676 | 14313680 | 4 | |
| Deletion | Chr1 | 14365158 | 14365160 | 2 | |
| Deletion | Chr5 | 15547178 | 15547179 | 1 | |
| Deletion | Chr8 | 16603860 | 16603862 | 2 | |
| Deletion | Chr11 | 17706272 | 17706275 | 3 | |
| Deletion | Chr4 | 18303803 | 18303804 | 1 | |
| Deletion | Chr2 | 19212627 | 19212634 | 7 | LOC_Os02g32480 |
| Deletion | Chr9 | 19861713 | 19861714 | 1 | |
| Deletion | Chr3 | 20142339 | 20142340 | 1 | |
| Deletion | Chr8 | 20613841 | 20613933 | 92 | |
| Deletion | Chr5 | 21687528 | 21687533 | 5 | LOC_Os05g37100 |
| Deletion | Chr7 | 24191205 | 24191206 | 1 | |
| Deletion | Chr8 | 25593042 | 25593043 | 1 | |
| Deletion | Chr5 | 29411022 | 29411023 | 1 | |
| Deletion | Chr1 | 32014869 | 32014874 | 5 | LOC_Os01g55570 |
| Deletion | Chr4 | 34057180 | 34057185 | 5 | |
| Deletion | Chr4 | 34077584 | 34077589 | 5 |
Insertions: 18
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 19800569 | 20588789 | |
| Inversion | Chr6 | 19800575 | 20588797 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 1706564 | Chr2 | 35082576 | |
| Translocation | Chr11 | 12909654 | Chr9 | 3993747 | |
| Translocation | Chr8 | 13239555 | Chr4 | 3609435 | LOC_Os04g06850 |
| Translocation | Chr8 | 16049363 | Chr4 | 13175658 | |
| Translocation | Chr11 | 24994393 | Chr4 | 17873076 |
