Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2922-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2922-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 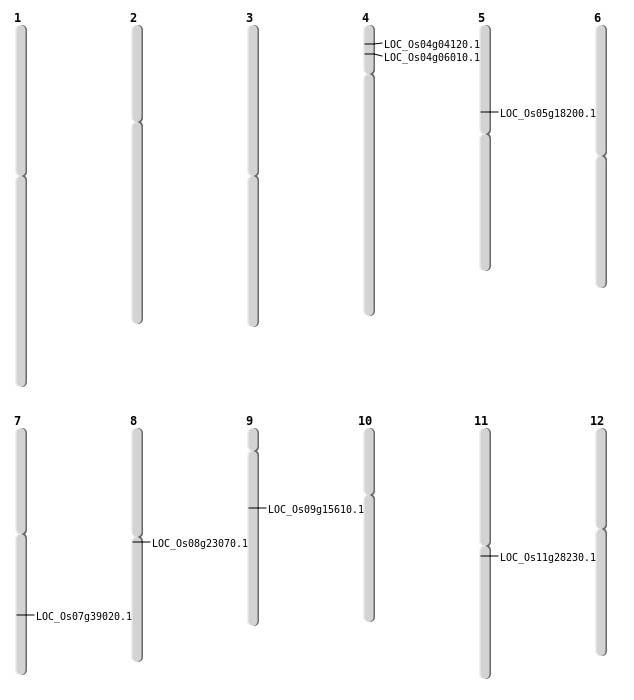
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20969627 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 25871166 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 29628266 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 6416883 | G-A | HET | INTRON | |
| SBS | Chr10 | 12654956 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 18213315 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 22309905 | G-C | HET | INTRON | |
| SBS | Chr11 | 16232331 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g28230 |
| SBS | Chr11 | 4145536 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 11468824 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 27052831 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 9480934 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 9481337 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 1886009 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 24393618 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 24435734 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 263294 | C-T | HET | START_GAINED | |
| SBS | Chr3 | 28977006 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 16916832 | A-G | HOMO | INTRON | |
| SBS | Chr4 | 17166970 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 19899232 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 3116779 | C-T | HET | STOP_GAINED | LOC_Os04g06010 |
| SBS | Chr4 | 5563269 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 28998351 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 17926305 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 25634274 | A-G | HOMO | INTRON | |
| SBS | Chr6 | 6968587 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 11220053 | G-A | HET | INTRON | |
| SBS | Chr7 | 11828990 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 15466031 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 21855712 | T-A | HET | INTRON | |
| SBS | Chr8 | 13772807 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 2216268 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 23667308 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 18819167 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 19218618 | T-A | HET | INTRON | |
| SBS | Chr9 | 8521780 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 9531147 | C-T | HET | SPLICE_SITE_DONOR | LOC_Os09g15610 |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 136674 | 136675 | 1 | |
| Deletion | Chr7 | 1494241 | 1494242 | 1 | |
| Deletion | Chr9 | 1621244 | 1621246 | 2 | |
| Deletion | Chr1 | 2998302 | 2998368 | 66 | |
| Deletion | Chr9 | 3908184 | 3908197 | 13 | |
| Deletion | Chr3 | 3968230 | 3968233 | 3 | |
| Deletion | Chr10 | 4063662 | 4063678 | 16 | |
| Deletion | Chr5 | 5012877 | 5012878 | 1 | |
| Deletion | Chr7 | 5038936 | 5038938 | 2 | |
| Deletion | Chr11 | 6458672 | 6458675 | 3 | |
| Deletion | Chr8 | 7005098 | 7005156 | 58 | |
| Deletion | Chr1 | 7362967 | 7362969 | 2 | |
| Deletion | Chr8 | 8223996 | 8224003 | 7 | |
| Deletion | Chr1 | 8469463 | 8469483 | 20 | |
| Deletion | Chr6 | 9012365 | 9012366 | 1 | |
| Deletion | Chr4 | 9331674 | 9331690 | 16 | |
| Deletion | Chr5 | 10472043 | 10472073 | 30 | LOC_Os05g18200 |
| Deletion | Chr1 | 10741728 | 10741733 | 5 | |
| Deletion | Chr1 | 10784994 | 10785063 | 69 | |
| Deletion | Chr7 | 12240311 | 12240313 | 2 | |
| Deletion | Chr12 | 12424124 | 12424126 | 2 | |
| Deletion | Chr5 | 12502764 | 12503567 | 803 | |
| Deletion | Chr1 | 13070839 | 13070840 | 1 | |
| Deletion | Chr8 | 13892776 | 13892788 | 12 | LOC_Os08g23070 |
| Deletion | Chr10 | 14073288 | 14073289 | 1 | |
| Deletion | Chr4 | 14628106 | 14628107 | 1 | |
| Deletion | Chr11 | 17706471 | 17706499 | 28 | |
| Deletion | Chr11 | 20126877 | 20126878 | 1 | |
| Deletion | Chr11 | 20236155 | 20236156 | 1 | |
| Deletion | Chr2 | 22191428 | 22191432 | 4 | |
| Deletion | Chr11 | 23645273 | 23645274 | 1 | |
| Deletion | Chr6 | 24605155 | 24605156 | 1 | |
| Deletion | Chr4 | 25520189 | 25520193 | 4 | |
| Deletion | Chr1 | 25737246 | 25737247 | 1 | |
| Deletion | Chr6 | 25802373 | 25802374 | 1 | |
| Deletion | Chr8 | 27473212 | 27473257 | 45 | |
| Deletion | Chr11 | 28616920 | 28616921 | 1 | |
| Deletion | Chr4 | 29879990 | 29880057 | 67 | |
| Deletion | Chr1 | 43256191 | 43256193 | 2 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 7910190 | 7910191 | 2 | |
| Insertion | Chr10 | 8384827 | 8384828 | 2 | |
| Insertion | Chr11 | 17184274 | 17184275 | 2 | |
| Insertion | Chr11 | 25836102 | 25836202 | 101 | |
| Insertion | Chr11 | 26729451 | 26729543 | 93 | |
| Insertion | Chr12 | 13048871 | 13048871 | 1 | |
| Insertion | Chr12 | 9852480 | 9852481 | 2 | |
| Insertion | Chr2 | 21645988 | 21645991 | 4 | |
| Insertion | Chr3 | 13517628 | 13517629 | 2 | |
| Insertion | Chr3 | 15789344 | 15789345 | 2 | |
| Insertion | Chr3 | 21554574 | 21554670 | 97 | |
| Insertion | Chr4 | 1898070 | 1898070 | 1 | LOC_Os04g04120 |
| Insertion | Chr4 | 2332987 | 2332987 | 1 | |
| Insertion | Chr4 | 7452431 | 7452431 | 1 | |
| Insertion | Chr5 | 11569354 | 11569356 | 3 | |
| Insertion | Chr5 | 19296501 | 19296607 | 107 | |
| Insertion | Chr6 | 11367496 | 11367496 | 1 | |
| Insertion | Chr9 | 6014066 | 6014141 | 76 | |
| Insertion | Chr9 | 94509 | 94510 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 11630753 | 12019471 | |
| Inversion | Chr6 | 11631521 | 12019472 | |
| Inversion | Chr7 | 23399647 | 23624849 | LOC_Os07g39020 |
| Inversion | Chr7 | 23399650 | 23624856 | LOC_Os07g39020 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 5238432 | Chr4 | 12976278 | |
| Translocation | Chr8 | 6341622 | Chr3 | 35852681 | |
| Translocation | Chr6 | 11630755 | Chr3 | 30845961 | |
| Translocation | Chr6 | 11631507 | Chr3 | 30845969 | |
| Translocation | Chr12 | 15592787 | Chr11 | 21832322 | |
| Translocation | Chr9 | 19071659 | Chr7 | 13538265 | |
| Translocation | Chr9 | 19071665 | Chr7 | 13346217 | |
| Translocation | Chr7 | 23767442 | Chr1 | 20739044 |
