Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2929-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2929-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 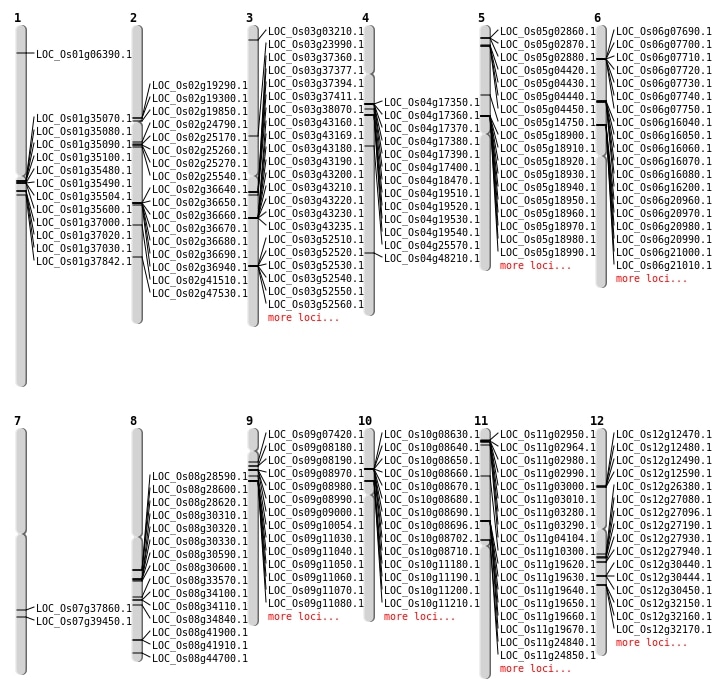
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16345631 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 406158 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 6524639 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 14454256 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 16892501 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 5384461 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 15511945 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 17678344 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 17989822 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 22893626 | G-T | HET | STOP_GAINED | LOC_Os12g37300 |
| SBS | Chr12 | 7743719 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 146247 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 146248 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 146249 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 16623606 | C-T | HET | INTRON | |
| SBS | Chr2 | 27476566 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 29039243 | C-T | HOMO | STOP_GAINED | LOC_Os02g47530 |
| SBS | Chr2 | 5455872 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 9074562 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 12944826 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 13615927 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g23990 |
| SBS | Chr3 | 17667892 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 21100918 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 21100919 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 25644115 | T-C | HET | INTRON | |
| SBS | Chr4 | 13158801 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 31725916 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11004363 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 6841476 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 11781747 | T-C | HET | INTRON | |
| SBS | Chr7 | 25312166 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 6624596 | A-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 1137588 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 15521857 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 8421144 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 16868978 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 16880025 | T-C | HET | SYNONYMOUS_CODING |
Deletions: 92
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 353144 | 353156 | 12 | |
| Deletion | Chr11 | 1020001 | 1047000 | 26999 | 6 |
| Deletion | Chr5 | 1029001 | 1047000 | 17999 | 4 |
| Deletion | Chr11 | 1203001 | 1231000 | 27999 | 3 |
| Deletion | Chr3 | 1364795 | 1364796 | 1 | LOC_Os03g03210 |
| Deletion | Chr5 | 2031001 | 2051000 | 19999 | 4 |
| Deletion | Chr3 | 2237470 | 2237472 | 2 | |
| Deletion | Chr4 | 2660788 | 2660789 | 1 | |
| Deletion | Chr4 | 2806199 | 2806203 | 4 | |
| Deletion | Chr1 | 3031000 | 3031013 | 13 | LOC_Os01g06390 |
| Deletion | Chr9 | 3591001 | 3603000 | 11999 | LOC_Os09g07420 |
| Deletion | Chr6 | 3715001 | 3757000 | 41999 | 7 |
| Deletion | Chr9 | 4236001 | 4253000 | 16999 | 2 |
| Deletion | Chr3 | 4618180 | 4618183 | 3 | |
| Deletion | Chr10 | 4680001 | 4732000 | 51999 | 10 |
| Deletion | Chr9 | 4763001 | 4799000 | 35999 | 5 |
| Deletion | Chr3 | 4783391 | 4783392 | 1 | |
| Deletion | Chr6 | 5874216 | 5874220 | 4 | |
| Deletion | Chr9 | 5932773 | 5932857 | 84 | |
| Deletion | Chr9 | 6121001 | 6172000 | 50999 | 13 |
| Deletion | Chr10 | 6190001 | 6219000 | 28999 | 4 |
| Deletion | Chr12 | 6861001 | 6886000 | 24999 | 4 |
| Deletion | Chr5 | 7548020 | 7548065 | 45 | |
| Deletion | Chr10 | 8074001 | 8088000 | 13999 | 2 |
| Deletion | Chr5 | 8238001 | 8252000 | 13999 | LOC_Os05g14750 |
| Deletion | Chr1 | 8347668 | 8347672 | 4 | |
| Deletion | Chr6 | 9119001 | 9156000 | 36999 | 6 |
| Deletion | Chr6 | 9293649 | 9293650 | 1 | |
| Deletion | Chr4 | 9503001 | 9520000 | 16999 | 7 |
| Deletion | Chr4 | 10862001 | 10886000 | 23999 | 4 |
| Deletion | Chr5 | 10971001 | 11021000 | 49999 | 10 |
| Deletion | Chr9 | 11017001 | 11055000 | 37999 | 2 |
| Deletion | Chr9 | 11097001 | 11110000 | 12999 | 3 |
| Deletion | Chr2 | 11267001 | 11282000 | 14999 | 3 |
| Deletion | Chr11 | 11307001 | 11330000 | 22999 | 6 |
| Deletion | Chr6 | 12115001 | 12149000 | 33999 | 8 |
| Deletion | Chr10 | 12136001 | 12149000 | 12999 | 2 |
| Deletion | Chr7 | 12260562 | 12260563 | 1 | |
| Deletion | Chr6 | 12559001 | 12570000 | 10999 | 2 |
| Deletion | Chr9 | 12853001 | 12863000 | 9999 | 2 |
| Deletion | Chr6 | 13429577 | 13429585 | 8 | |
| Deletion | Chr6 | 13527335 | 13527337 | 2 | |
| Deletion | Chr6 | 14122001 | 14152000 | 29999 | 4 |
| Deletion | Chr11 | 14156001 | 14246000 | 89999 | 18 |
| Deletion | Chr2 | 14373001 | 14386000 | 12999 | 2 |
| Deletion | Chr3 | 14459260 | 14459268 | 8 | |
| Deletion | Chr2 | 14704001 | 14723000 | 18999 | 3 |
| Deletion | Chr3 | 14947373 | 14947378 | 5 | |
| Deletion | Chr12 | 15405001 | 15415000 | 9999 | 2 |
| Deletion | Chr5 | 15518001 | 15535000 | 16999 | 4 |
| Deletion | Chr4 | 15697810 | 15697811 | 1 | |
| Deletion | Chr6 | 15806001 | 15825000 | 18999 | 2 |
| Deletion | Chr12 | 15864001 | 15893000 | 28999 | 2 |
| Deletion | Chr12 | 16420229 | 16420237 | 8 | |
| Deletion | Chr12 | 16457001 | 16472000 | 14999 | 2 |
| Deletion | Chr6 | 17029001 | 17067000 | 37999 | 5 |
| Deletion | Chr6 | 17317001 | 17339000 | 21999 | 3 |
| Deletion | Chr8 | 17457001 | 17479000 | 21999 | 3 |
| Deletion | Chr1 | 17597699 | 17597707 | 8 | |
| Deletion | Chr12 | 18257001 | 18279000 | 21999 | 3 |
| Deletion | Chr8 | 18652001 | 18675000 | 22999 | 3 |
| Deletion | Chr8 | 18843001 | 18863000 | 19999 | 2 |
| Deletion | Chr12 | 19384001 | 19420000 | 35999 | 4 |
| Deletion | Chr1 | 19408001 | 19436000 | 27999 | 4 |
| Deletion | Chr5 | 19553001 | 19575000 | 21999 | 2 |
| Deletion | Chr1 | 19628001 | 19648000 | 19999 | 4 |
| Deletion | Chr1 | 20636001 | 20662000 | 25999 | 4 |
| Deletion | Chr3 | 20699001 | 20737000 | 37999 | 5 |
| Deletion | Chr9 | 20916001 | 20936000 | 19999 | 2 |
| Deletion | Chr8 | 21370001 | 21392000 | 21999 | 3 |
| Deletion | Chr12 | 21735001 | 21768000 | 32999 | 4 |
| Deletion | Chr2 | 22114001 | 22146000 | 31999 | 7 |
| Deletion | Chr7 | 22695001 | 22707000 | 11999 | 2 |
| Deletion | Chr3 | 24076001 | 24111000 | 34999 | 9 |
| Deletion | Chr2 | 24846001 | 24859000 | 12999 | LOC_Os02g41510 |
| Deletion | Chr2 | 25112047 | 25112064 | 17 | |
| Deletion | Chr2 | 25167600 | 25167601 | 1 | |
| Deletion | Chr6 | 25377846 | 25377847 | 1 | |
| Deletion | Chr3 | 25385053 | 25385054 | 1 | |
| Deletion | Chr7 | 25901536 | 25901543 | 7 | |
| Deletion | Chr5 | 26102001 | 26149000 | 46999 | 7 |
| Deletion | Chr8 | 26456001 | 26481000 | 24999 | 2 |
| Deletion | Chr11 | 26620001 | 26644000 | 23999 | 7 |
| Deletion | Chr4 | 28378001 | 28389000 | 10999 | LOC_Os04g48210 |
| Deletion | Chr1 | 28807960 | 28807969 | 9 | |
| Deletion | Chr6 | 29260603 | 29260604 | 1 | |
| Deletion | Chr5 | 29956352 | 29956354 | 2 | |
| Deletion | Chr3 | 30125001 | 30175000 | 49999 | 8 |
| Deletion | Chr1 | 32286857 | 32286861 | 4 | |
| Deletion | Chr2 | 32469048 | 32469056 | 8 | |
| Deletion | Chr1 | 40789597 | 40789598 | 1 | |
| Deletion | Chr1 | 41084381 | 41084386 | 5 |
Insertions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 8241519 | 8241520 | 2 | |
| Insertion | Chr10 | 6227267 | 6227296 | 30 | |
| Insertion | Chr11 | 1032498 | 1032498 | 1 | |
| Insertion | Chr11 | 5592439 | 5592439 | 1 | LOC_Os11g10300 |
| Insertion | Chr3 | 11157164 | 11157165 | 2 | |
| Insertion | Chr3 | 1368044 | 1368044 | 1 | LOC_Os03g03210 |
| Insertion | Chr3 | 17832414 | 17832420 | 7 | |
| Insertion | Chr4 | 10383840 | 10383840 | 1 | |
| Insertion | Chr4 | 13544345 | 13544345 | 1 | |
| Insertion | Chr4 | 14826498 | 14826617 | 120 | LOC_Os04g25570 |
| Insertion | Chr4 | 2858320 | 2858321 | 2 | |
| Insertion | Chr6 | 10048768 | 10048769 | 2 | |
| Insertion | Chr7 | 14384250 | 14384282 | 33 | |
| Insertion | Chr7 | 14843846 | 14843846 | 1 | |
| Insertion | Chr7 | 9373879 | 9373880 | 2 | |
| Insertion | Chr8 | 16458425 | 16458438 | 14 | |
| Insertion | Chr8 | 16650817 | 16650817 | 1 | |
| Insertion | Chr8 | 20962475 | 20962475 | 1 | LOC_Os08g33570 |
| Insertion | Chr8 | 28089917 | 28089917 | 1 | LOC_Os08g44700 |
| Insertion | Chr9 | 11420116 | 11420227 | 112 | |
| Insertion | Chr9 | 22463082 | 22463093 | 12 | |
| Insertion | Chr9 | 3159849 | 3159851 | 3 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 12004788 | 12005556 | |
| Inversion | Chr4 | 23501980 | 23504495 | |
| Inversion | Chr4 | 23502007 | 23504502 |
No Translocation
