Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2940-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2940-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 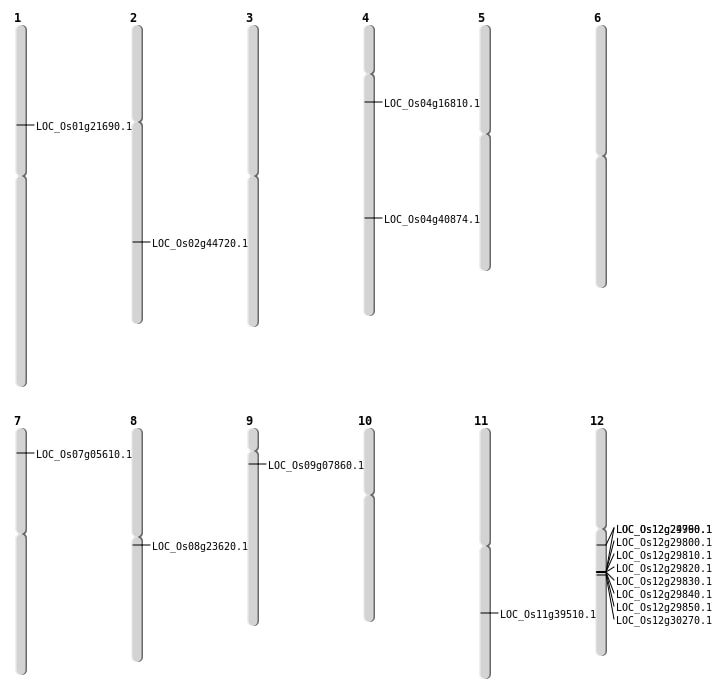
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12201198 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 15596932 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 3635517 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 18838613 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 25183854 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 11495046 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11871698 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14330502 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g24960 |
| SBS | Chr12 | 17873062 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 24043240 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 9031833 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 9031842 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 14623132 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 1541871 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 17270623 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 18227078 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 21816983 | C-T | HET | INTRON | |
| SBS | Chr3 | 30157618 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 8358049 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 19965044 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 24717863 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 31671858 | T-C | HOMO | INTRON | |
| SBS | Chr4 | 9212804 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16810 |
| SBS | Chr5 | 12804950 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 17197849 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr5 | 19628689 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 7855170 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 942302 | G-A | HET | INTRON | |
| SBS | Chr7 | 12403177 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22365025 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 2635488 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g05610 |
| SBS | Chr8 | 1078981 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 14299832 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23620 |
| SBS | Chr8 | 17326338 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 6224330 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 21812197 | A-G | HET | INTRON | |
| SBS | Chr9 | 21812198 | A-G | HET | INTRON | |
| SBS | Chr9 | 3997712 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 107234 | 107236 | 2 | |
| Deletion | Chr9 | 130379 | 130380 | 1 | |
| Deletion | Chr4 | 535983 | 535992 | 9 | |
| Deletion | Chr8 | 603684 | 603685 | 1 | |
| Deletion | Chr2 | 3667280 | 3667290 | 10 | |
| Deletion | Chr9 | 5730508 | 5730509 | 1 | |
| Deletion | Chr1 | 6161511 | 6161512 | 1 | |
| Deletion | Chr7 | 6209039 | 6209040 | 1 | |
| Deletion | Chr7 | 6798318 | 6798319 | 1 | |
| Deletion | Chr6 | 8610052 | 8610053 | 1 | |
| Deletion | Chr12 | 9118198 | 9118199 | 1 | |
| Deletion | Chr7 | 9590959 | 9590960 | 1 | |
| Deletion | Chr10 | 9732795 | 9732796 | 1 | |
| Deletion | Chr9 | 9851225 | 9851226 | 1 | |
| Deletion | Chr9 | 10594467 | 10594469 | 2 | |
| Deletion | Chr6 | 11088113 | 11088114 | 1 | |
| Deletion | Chr1 | 12168241 | 12168243 | 2 | LOC_Os01g21690 |
| Deletion | Chr12 | 12299692 | 12299693 | 1 | |
| Deletion | Chr8 | 12812744 | 12812755 | 11 | |
| Deletion | Chr2 | 13698430 | 13698751 | 321 | |
| Deletion | Chr12 | 17798001 | 17853000 | 54999 | 8 |
| Deletion | Chr2 | 18782842 | 18782879 | 37 | |
| Deletion | Chr10 | 20077856 | 20077857 | 1 | |
| Deletion | Chr5 | 20267338 | 20267350 | 12 | |
| Deletion | Chr11 | 21126278 | 21126280 | 2 | |
| Deletion | Chr11 | 24787732 | 24787755 | 23 | |
| Deletion | Chr2 | 24878001 | 24887000 | 8999 | LOC_Os02g44720 |
| Deletion | Chr3 | 28218060 | 28218061 | 1 | |
| Deletion | Chr6 | 29550967 | 29550968 | 1 | |
| Deletion | Chr1 | 31730768 | 31730769 | 1 |
Insertions: 16
Inversions: 7
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 985230 | Chr1 | 25907714 | |
| Translocation | Chr8 | 6088991 | Chr3 | 3629975 | |
| Translocation | Chr8 | 6089753 | Chr3 | 3629966 | |
| Translocation | Chr5 | 12223018 | Chr1 | 40255502 | |
| Translocation | Chr5 | 12223344 | Chr1 | 27211624 | |
| Translocation | Chr11 | 23519623 | Chr4 | 24265986 | LOC_Os04g40874 |
| Translocation | Chr11 | 23522693 | Chr4 | 24270555 | LOC_Os11g39510 |
| Translocation | Chr11 | 25115600 | Chr5 | 26317109 |
