Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2942-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2942-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 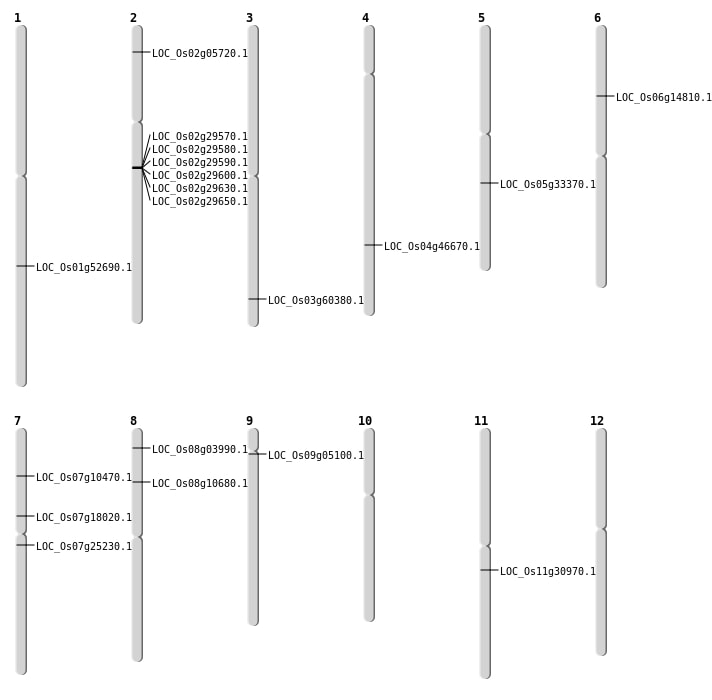
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28071529 | C-A | HOMO | INTRON | |
| SBS | Chr10 | 23172769 | T-C | HET | INTRON | |
| SBS | Chr11 | 16079537 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 18018371 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g30970 |
| SBS | Chr11 | 23606374 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 11036332 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11519557 | A-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 21122262 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 5695033 | T-C | HOMO | INTRON | |
| SBS | Chr2 | 12289667 | C-T | HET | INTRON | |
| SBS | Chr2 | 25674067 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr2 | 27231296 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 216024 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 35301574 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 13532525 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 26598603 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 27667028 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g46670 |
| SBS | Chr4 | 28510509 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 34225230 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 1599817 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13741434 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 6247525 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr6 | 9107566 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 10032723 | T-A | HET | INTRON | |
| SBS | Chr7 | 10607278 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 78110 | A-T | HET | INTRON | |
| SBS | Chr8 | 10892907 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 21841117 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 109832 | A-T | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1916845 | 1916852 | 7 | LOC_Os08g03990 |
| Deletion | Chr2 | 2814602 | 2814603 | 1 | LOC_Os02g05720 |
| Deletion | Chr10 | 3145218 | 3145219 | 1 | |
| Deletion | Chr6 | 3762508 | 3762514 | 6 | |
| Deletion | Chr11 | 3986114 | 3986115 | 1 | |
| Deletion | Chr5 | 4446139 | 4446140 | 1 | |
| Deletion | Chr6 | 5065652 | 5065660 | 8 | |
| Deletion | Chr7 | 5633736 | 5633737 | 1 | LOC_Os07g10470 |
| Deletion | Chr8 | 6294268 | 6294269 | 1 | LOC_Os08g10680 |
| Deletion | Chr4 | 6598668 | 6598669 | 1 | |
| Deletion | Chr11 | 7731082 | 7731096 | 14 | |
| Deletion | Chr6 | 8410048 | 8410049 | 1 | LOC_Os06g14810 |
| Deletion | Chr8 | 9279448 | 9279449 | 1 | |
| Deletion | Chr12 | 9615535 | 9615628 | 93 | |
| Deletion | Chr3 | 12759629 | 12759656 | 27 | |
| Deletion | Chr12 | 16890436 | 16890438 | 2 | |
| Deletion | Chr10 | 17392605 | 17392628 | 23 | |
| Deletion | Chr2 | 17567001 | 17612000 | 44999 | 5 |
| Deletion | Chr5 | 19603853 | 19603858 | 5 | LOC_Os05g33370 |
| Deletion | Chr12 | 21152828 | 21152832 | 4 | |
| Deletion | Chr3 | 23681838 | 23681839 | 1 | |
| Deletion | Chr2 | 26131738 | 26131744 | 6 | |
| Deletion | Chr7 | 27252376 | 27252386 | 10 | |
| Deletion | Chr11 | 27615479 | 27615489 | 10 | |
| Deletion | Chr4 | 28104062 | 28104151 | 89 | |
| Deletion | Chr2 | 29460847 | 29460854 | 7 | |
| Deletion | Chr4 | 29699250 | 29699251 | 1 | |
| Deletion | Chr1 | 30295519 | 30295532 | 13 | LOC_Os01g52690 |
| Deletion | Chr2 | 34528819 | 34528823 | 4 | |
| Deletion | Chr1 | 37704058 | 37704108 | 50 |
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 31479258 | 31479259 | 2 | |
| Insertion | Chr10 | 22789987 | 22790007 | 21 | |
| Insertion | Chr10 | 6366909 | 6366910 | 2 | |
| Insertion | Chr11 | 26982523 | 26982523 | 1 | |
| Insertion | Chr3 | 8825062 | 8825070 | 9 | |
| Insertion | Chr3 | 9059126 | 9059128 | 3 | |
| Insertion | Chr4 | 7871912 | 7871934 | 23 | |
| Insertion | Chr5 | 11149272 | 11149272 | 1 | |
| Insertion | Chr6 | 1246778 | 1246804 | 27 | |
| Insertion | Chr7 | 13708741 | 13708741 | 1 | |
| Insertion | Chr7 | 14422450 | 14422450 | 1 | LOC_Os07g25230 |
| Insertion | Chr7 | 20331374 | 20331374 | 1 | |
| Insertion | Chr8 | 144604 | 144604 | 1 | |
| Insertion | Chr8 | 17291236 | 17291236 | 1 | |
| Insertion | Chr8 | 3876548 | 3876548 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 15845491 | 15847588 | |
| Inversion | Chr2 | 17568190 | 18006530 | |
| Inversion | Chr2 | 17622719 | 18006537 | LOC_Os02g29630 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 2741905 | Chr7 | 10663580 | 2 |
| Translocation | Chr4 | 12290058 | Chr3 | 34340975 | LOC_Os03g60380 |
| Translocation | Chr7 | 18542654 | Chr4 | 17812135 |
