Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2947-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2947-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 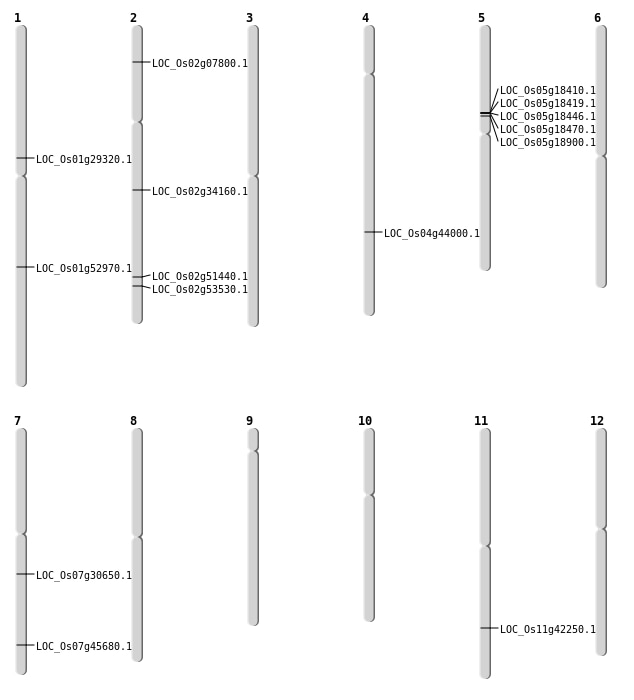
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 3744876 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 11938498 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 14707325 | C-T | HET | INTRON | |
| SBS | Chr11 | 18222863 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 24140215 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 25455408 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g42250 |
| SBS | Chr12 | 14068105 | G-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 195570 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 20429320 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34160 |
| SBS | Chr2 | 30193443 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 31773576 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 32750792 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g53530 |
| SBS | Chr2 | 34475701 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 68919 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 11651336 | A-G | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 12666729 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 7992392 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 20942518 | A-C | HOMO | INTRON | |
| SBS | Chr4 | 25430261 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 33511067 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 7481375 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 20914055 | G-A | HET | INTRON | |
| SBS | Chr5 | 22291931 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 24778215 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 1449528 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 16536956 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 19059338 | T-C | HOMO | INTRON | |
| SBS | Chr7 | 27275496 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45680 |
| SBS | Chr8 | 10030938 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 10539215 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 10890453 | G-A | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 4052874 | 4052876 | 2 | |
| Deletion | Chr2 | 4083385 | 4083389 | 4 | LOC_Os02g07800 |
| Deletion | Chr12 | 5011736 | 5011829 | 93 | |
| Deletion | Chr9 | 7115595 | 7115692 | 97 | |
| Deletion | Chr2 | 8123449 | 8123457 | 8 | |
| Deletion | Chr2 | 9242777 | 9242779 | 2 | |
| Deletion | Chr5 | 10636001 | 10674000 | 37999 | 5 |
| Deletion | Chr12 | 14064739 | 14064741 | 2 | |
| Deletion | Chr3 | 15300188 | 15300198 | 10 | |
| Deletion | Chr8 | 15762409 | 15762416 | 7 | |
| Deletion | Chr7 | 15934295 | 15934296 | 1 | |
| Deletion | Chr1 | 16427628 | 16427646 | 18 | LOC_Os01g29320 |
| Deletion | Chr5 | 17136987 | 17136991 | 4 | |
| Deletion | Chr7 | 18139912 | 18139914 | 2 | LOC_Os07g30650 |
| Deletion | Chr1 | 18477918 | 18477936 | 18 | |
| Deletion | Chr7 | 19491521 | 19491532 | 11 | |
| Deletion | Chr3 | 19977899 | 19977910 | 11 | |
| Deletion | Chr7 | 20469627 | 20469631 | 4 | |
| Deletion | Chr6 | 21780344 | 21780365 | 21 | |
| Deletion | Chr10 | 21785205 | 21785216 | 11 | |
| Deletion | Chr4 | 22612001 | 22612010 | 9 | |
| Deletion | Chr2 | 22702097 | 22702099 | 2 | |
| Deletion | Chr4 | 26083643 | 26083644 | 1 | LOC_Os04g44000 |
| Deletion | Chr12 | 27236984 | 27236985 | 1 | |
| Deletion | Chr1 | 29212171 | 29212172 | 1 | |
| Deletion | Chr1 | 30447645 | 30447659 | 14 | LOC_Os01g52970 |
| Deletion | Chr3 | 31180263 | 31180287 | 24 |
Insertions: 8
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 16209412 | 16586979 | |
| Inversion | Chr8 | 16209419 | 16586980 | |
| Inversion | Chr3 | 26098372 | 26196038 | |
| Inversion | Chr3 | 26098376 | 26196037 | |
| Inversion | Chr3 | 26463488 | 27191784 | |
| Inversion | Chr3 | 26463498 | 27191796 | |
| Inversion | Chr2 | 31352121 | 31517552 | |
| Inversion | Chr2 | 31352131 | 31517659 | LOC_Os02g51440 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 10659256 | Chr6 | 15618050 | |
| Translocation | Chr12 | 13352820 | Chr1 | 18686327 |
