Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2951-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2951-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 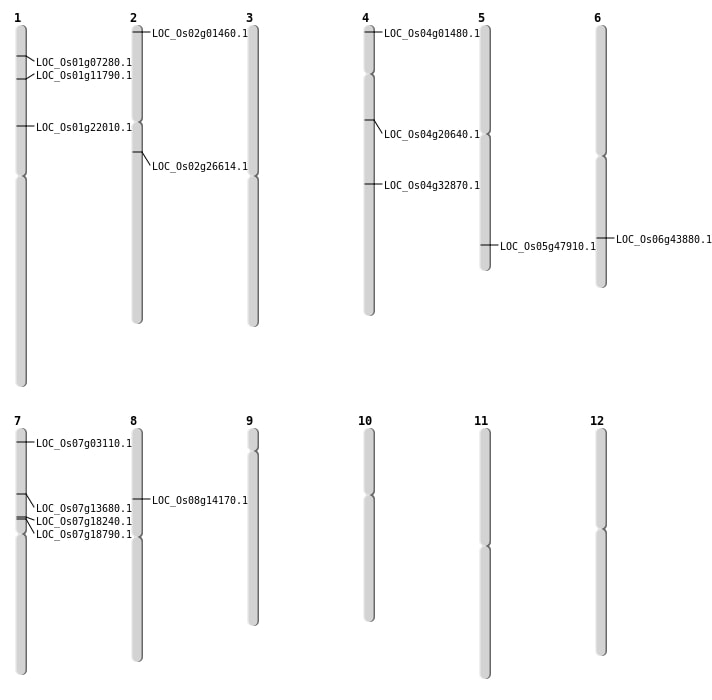
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24974132 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 3433747 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g07280 |
| SBS | Chr1 | 5932512 | C-A | HET | INTRON | |
| SBS | Chr10 | 12427593 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 22358169 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 22358170 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 3192187 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 6617679 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 13052102 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 8801887 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 16600706 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 15626683 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g26614 |
| SBS | Chr2 | 15911363 | G-A | HET | INTRON | |
| SBS | Chr2 | 27088884 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 35271652 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr2 | 35271654 | G-T | HET | START_GAINED | |
| SBS | Chr2 | 6690898 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 29166787 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 11553475 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g20640 |
| SBS | Chr4 | 18395644 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 321717 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g01480 |
| SBS | Chr6 | 14041752 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 24370798 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 26238325 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 7339143 | T-C | HET | INTRON | |
| SBS | Chr7 | 11121085 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18790 |
| SBS | Chr7 | 14225238 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 7853027 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13680 |
| SBS | Chr8 | 8460526 | C-T | HET | STOP_GAINED | LOC_Os08g14170 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 270161 | 270170 | 9 | LOC_Os02g01460 |
| Deletion | Chr7 | 1185382 | 1185397 | 15 | LOC_Os07g03110 |
| Deletion | Chr7 | 1409279 | 1409283 | 4 | |
| Deletion | Chr9 | 3292717 | 3292718 | 1 | |
| Deletion | Chr4 | 4853057 | 4853143 | 86 | |
| Deletion | Chr1 | 6381738 | 6381742 | 4 | LOC_Os01g11790 |
| Deletion | Chr6 | 7029076 | 7029078 | 2 | |
| Deletion | Chr1 | 8177047 | 8177049 | 2 | |
| Deletion | Chr5 | 8759769 | 8759778 | 9 | |
| Deletion | Chr3 | 11436983 | 11436984 | 1 | |
| Deletion | Chr5 | 12942218 | 12942230 | 12 | |
| Deletion | Chr7 | 13632907 | 13632908 | 1 | |
| Deletion | Chr11 | 15724983 | 15724991 | 8 | |
| Deletion | Chr1 | 16855373 | 16855374 | 1 | |
| Deletion | Chr4 | 19849657 | 19849661 | 4 | LOC_Os04g32870 |
| Deletion | Chr2 | 19890360 | 19890402 | 42 | |
| Deletion | Chr12 | 20326729 | 20326730 | 1 | |
| Deletion | Chr10 | 23083175 | 23083180 | 5 | |
| Deletion | Chr4 | 23640298 | 23640304 | 6 | |
| Deletion | Chr3 | 23921453 | 23921471 | 18 | |
| Deletion | Chr3 | 24007635 | 24007636 | 1 | |
| Deletion | Chr7 | 24223393 | 24223403 | 10 | |
| Deletion | Chr1 | 25822942 | 25822990 | 48 | |
| Deletion | Chr4 | 26058546 | 26058552 | 6 | |
| Deletion | Chr6 | 26435941 | 26435942 | 1 | LOC_Os06g43880 |
| Deletion | Chr12 | 26686172 | 26686182 | 10 | |
| Deletion | Chr5 | 27466641 | 27466649 | 8 | LOC_Os05g47910 |
| Deletion | Chr4 | 28032467 | 28032468 | 1 |
Insertions: 5
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 3853850 | 3912700 | |
| Inversion | Chr3 | 3853867 | 3912703 | |
| Inversion | Chr1 | 12366264 | 12512344 | LOC_Os01g22010 |
| Inversion | Chr4 | 16096371 | 16494242 | |
| Inversion | Chr3 | 25369655 | 25835250 | |
| Inversion | Chr3 | 25814853 | 25835245 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 10812725 | Chr6 | 24395506 | LOC_Os07g18240 |
| Translocation | Chr6 | 19238242 | Chr4 | 11584971 | |
| Translocation | Chr10 | 22358960 | Chr6 | 28454356 |
