Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2968-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2968-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 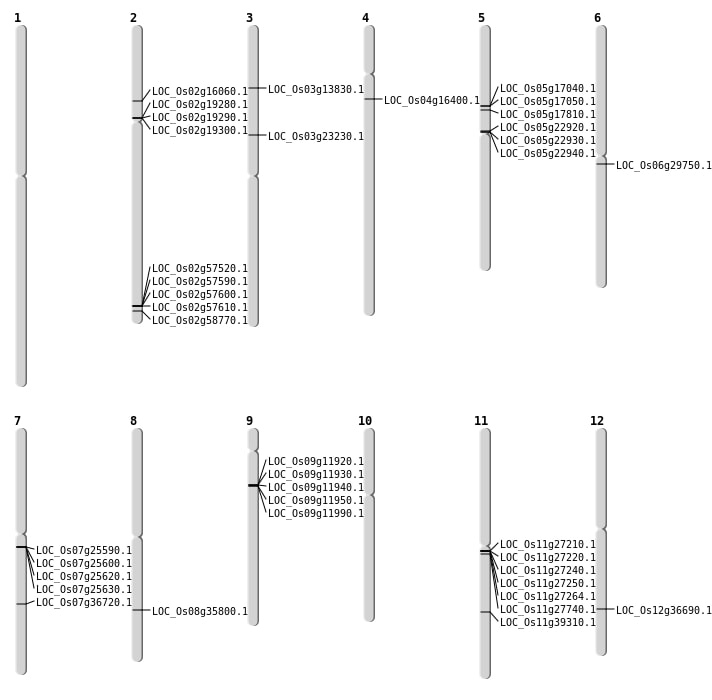
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11018735 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 16211972 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 19218323 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3240263 | C-T | HET | INTRON | |
| SBS | Chr10 | 4473759 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 14820696 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 15096452 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22477819 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g36690 |
| SBS | Chr12 | 22477820 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 7410190 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 18695949 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 19632397 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 20036548 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 21185472 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 17541647 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 20310418 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 20310419 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28478290 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 35851135 | C-T | HET | INTRON | |
| SBS | Chr3 | 7498512 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g13830 |
| SBS | Chr4 | 8909004 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16400 |
| SBS | Chr5 | 22883234 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 132463 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 21998655 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g36720 |
| SBS | Chr7 | 7202681 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 22577877 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g35800 |
| SBS | Chr8 | 22645991 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 27887799 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 9091813 | A-T | HET | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1527992 | 1528082 | 90 | |
| Deletion | Chr12 | 4419501 | 4419512 | 11 | |
| Deletion | Chr4 | 5293511 | 5293512 | 1 | |
| Deletion | Chr12 | 5447822 | 5447823 | 1 | |
| Deletion | Chr9 | 5560396 | 5560397 | 1 | |
| Deletion | Chr9 | 6694001 | 6737000 | 42999 | 5 |
| Deletion | Chr8 | 6771211 | 6771212 | 1 | |
| Deletion | Chr4 | 7655009 | 7655011 | 2 | |
| Deletion | Chr7 | 8694006 | 8694007 | 1 | |
| Deletion | Chr5 | 9717001 | 9743000 | 25999 | 3 |
| Deletion | Chr11 | 11222437 | 11222439 | 2 | |
| Deletion | Chr2 | 11261001 | 11282000 | 20999 | 3 |
| Deletion | Chr7 | 11802472 | 11802476 | 4 | |
| Deletion | Chr1 | 12473427 | 12473431 | 4 | |
| Deletion | Chr5 | 13040001 | 13072000 | 31999 | 3 |
| Deletion | Chr10 | 13314422 | 13314435 | 13 | |
| Deletion | Chr3 | 13447888 | 13447923 | 35 | LOC_Os03g23230 |
| Deletion | Chr7 | 14674001 | 14698000 | 23999 | 4 |
| Deletion | Chr5 | 15397960 | 15397961 | 1 | |
| Deletion | Chr11 | 15646001 | 15693000 | 46999 | 6 |
| Deletion | Chr11 | 15770251 | 15770299 | 48 | |
| Deletion | Chr12 | 16443505 | 16443507 | 2 | |
| Deletion | Chr1 | 16657119 | 16657121 | 2 | |
| Deletion | Chr10 | 17062669 | 17062670 | 1 | |
| Deletion | Chr6 | 17084368 | 17084419 | 51 | LOC_Os06g29750 |
| Deletion | Chr8 | 18395712 | 18395713 | 1 | |
| Deletion | Chr7 | 20943388 | 20943394 | 6 | |
| Deletion | Chr6 | 22482944 | 22482954 | 10 | |
| Deletion | Chr5 | 27816043 | 27816071 | 28 | |
| Deletion | Chr2 | 30163860 | 30163870 | 10 | |
| Deletion | Chr1 | 34239790 | 34239791 | 1 | |
| Deletion | Chr2 | 35270001 | 35284000 | 13999 | 4 |
Insertions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 15482047 | 15482048 | 2 | |
| Insertion | Chr12 | 20737222 | 20737222 | 1 | |
| Insertion | Chr12 | 25500420 | 25500420 | 1 | |
| Insertion | Chr12 | 9191945 | 9191945 | 1 | |
| Insertion | Chr2 | 15267660 | 15267661 | 2 | |
| Insertion | Chr2 | 22286518 | 22286519 | 2 | |
| Insertion | Chr2 | 9139264 | 9139264 | 1 | LOC_Os02g16060 |
| Insertion | Chr3 | 11687381 | 11687414 | 34 | |
| Insertion | Chr3 | 8952516 | 8952517 | 2 | |
| Insertion | Chr4 | 1058037 | 1058037 | 1 | |
| Insertion | Chr4 | 2655878 | 2655880 | 3 | |
| Insertion | Chr6 | 11923167 | 11923167 | 1 | |
| Insertion | Chr6 | 25551052 | 25551053 | 2 | |
| Insertion | Chr6 | 4597882 | 4597883 | 2 | |
| Insertion | Chr7 | 21202638 | 21202641 | 4 | |
| Insertion | Chr7 | 2658155 | 2658258 | 104 | |
| Insertion | Chr7 | 27613274 | 27613274 | 1 | |
| Insertion | Chr8 | 17284959 | 17284959 | 1 | |
| Insertion | Chr8 | 26825109 | 26825169 | 61 | |
| Insertion | Chr9 | 21645029 | 21645029 | 1 |
No Inversion
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 2747255 | Chr5 | 977146 | |
| Translocation | Chr10 | 9175607 | Chr3 | 22377469 | |
| Translocation | Chr10 | 9175612 | Chr3 | 22377049 | |
| Translocation | Chr11 | 11221807 | Chr2 | 35243787 | LOC_Os02g57520 |
| Translocation | Chr4 | 11231489 | Chr3 | 4244405 | |
| Translocation | Chr12 | 18487858 | Chr11 | 18175911 | |
| Translocation | Chr11 | 23404412 | Chr2 | 35266784 | LOC_Os11g39310 |
| Translocation | Chr11 | 23404417 | Chr3 | 34653225 | LOC_Os11g39310 |
| Translocation | Chr11 | 23404730 | Chr3 | 34653219 | LOC_Os11g39310 |
| Translocation | Chr8 | 26997820 | Chr5 | 23804825 |
