Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2970-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2970-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 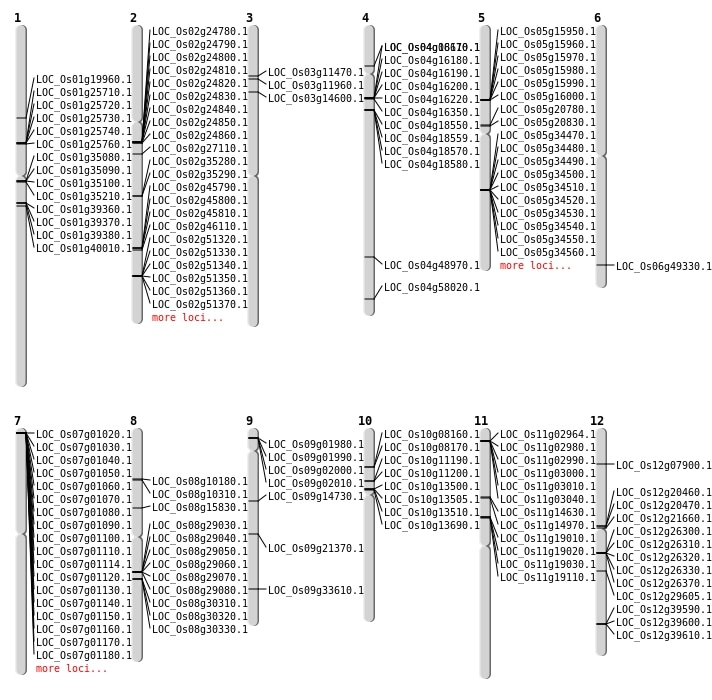
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21442377 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 39410801 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 39410806 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 40490384 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 13810701 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 15535465 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18735167 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 6353123 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 6353124 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 6353125 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 8509111 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 14785757 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 16080801 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 19067207 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 28489227 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 29306489 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 18105228 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 20601228 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 20747778 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 27454431 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 9773590 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 17670352 | A-C | HOMO | INTRON | |
| SBS | Chr4 | 2013827 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 34558967 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g58020 |
| SBS | Chr4 | 35479638 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 4682026 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g08610 |
| SBS | Chr5 | 143975 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr5 | 18627717 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20591241 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 1243518 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 7544400 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 20306376 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 9030918 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 12899378 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g21370 |
| SBS | Chr9 | 12939582 | A-G | HOMO | INTRON | |
| SBS | Chr9 | 17277020 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 19847871 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 19847872 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g33610 |
| SBS | Chr9 | 20927537 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 8752526 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14730 |
| SBS | Chr9 | 9840475 | T-G | HET | INTERGENIC |
Deletions: 69
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1 | 845000 | 844999 | 138 |
| Deletion | Chr7 | 545670 | 545673 | 3 | |
| Deletion | Chr9 | 682001 | 702000 | 19999 | 4 |
| Deletion | Chr11 | 1023001 | 1049000 | 25999 | 6 |
| Deletion | Chr1 | 3951643 | 3951645 | 2 | |
| Deletion | Chr4 | 4187104 | 4187108 | 4 | |
| Deletion | Chr10 | 4398001 | 4416000 | 17999 | 2 |
| Deletion | Chr9 | 4753131 | 4753204 | 73 | |
| Deletion | Chr9 | 4904955 | 4904963 | 8 | |
| Deletion | Chr4 | 5019177 | 5019192 | 15 | |
| Deletion | Chr8 | 5896001 | 5907000 | 10999 | 2 |
| Deletion | Chr3 | 5913001 | 5928000 | 14999 | 2 |
| Deletion | Chr10 | 6197001 | 6213000 | 15999 | 2 |
| Deletion | Chr5 | 6821435 | 6821436 | 1 | |
| Deletion | Chr10 | 7286001 | 7317000 | 30999 | 4 |
| Deletion | Chr8 | 7414078 | 7414079 | 1 | |
| Deletion | Chr2 | 7906071 | 7906072 | 1 | |
| Deletion | Chr3 | 7925608 | 7925623 | 15 | LOC_Os03g14600 |
| Deletion | Chr11 | 8142004 | 8142007 | 3 | |
| Deletion | Chr11 | 8145616 | 8145641 | 25 | |
| Deletion | Chr11 | 8237001 | 8247000 | 9999 | 2 |
| Deletion | Chr8 | 8545561 | 8545562 | 1 | |
| Deletion | Chr11 | 8588766 | 8588767 | 1 | |
| Deletion | Chr4 | 8778001 | 8812000 | 33999 | 6 |
| Deletion | Chr5 | 9004001 | 9036000 | 31999 | 6 |
| Deletion | Chr8 | 9064370 | 9064379 | 9 | |
| Deletion | Chr9 | 9097561 | 9097570 | 9 | |
| Deletion | Chr8 | 9621855 | 9621858 | 3 | LOC_Os08g15830 |
| Deletion | Chr2 | 10073741 | 10073742 | 1 | |
| Deletion | Chr9 | 10082712 | 10082720 | 8 | |
| Deletion | Chr4 | 10243001 | 10265000 | 21999 | 4 |
| Deletion | Chr11 | 10833001 | 10853000 | 19999 | 4 |
| Deletion | Chr5 | 10936804 | 10936805 | 1 | |
| Deletion | Chr4 | 11182846 | 11182860 | 14 | |
| Deletion | Chr11 | 11217337 | 11217341 | 4 | |
| Deletion | Chr1 | 11333649 | 11333652 | 3 | LOC_Os01g19960 |
| Deletion | Chr10 | 11456298 | 11456359 | 61 | |
| Deletion | Chr12 | 11616289 | 11616290 | 1 | |
| Deletion | Chr12 | 11942001 | 11966000 | 23999 | 3 |
| Deletion | Chr10 | 12034434 | 12034441 | 7 | |
| Deletion | Chr5 | 12189001 | 12202000 | 12999 | 2 |
| Deletion | Chr9 | 14215712 | 14215713 | 1 | |
| Deletion | Chr2 | 14368001 | 14415000 | 46999 | 9 |
| Deletion | Chr1 | 14574001 | 14600000 | 25999 | 5 |
| Deletion | Chr12 | 15230814 | 15230818 | 4 | |
| Deletion | Chr12 | 15334001 | 15385000 | 50999 | 5 |
| Deletion | Chr2 | 15932001 | 15945000 | 12999 | LOC_Os02g27110 |
| Deletion | Chr10 | 16014368 | 16014369 | 1 | |
| Deletion | Chr5 | 16923169 | 16923181 | 12 | |
| Deletion | Chr6 | 17068260 | 17068270 | 10 | |
| Deletion | Chr12 | 17661072 | 17661073 | 1 | LOC_Os12g29605 |
| Deletion | Chr8 | 17771001 | 17812000 | 40999 | 6 |
| Deletion | Chr8 | 18654001 | 18671000 | 16999 | 3 |
| Deletion | Chr1 | 19415001 | 19436000 | 20999 | 4 |
| Deletion | Chr10 | 19877159 | 19877166 | 7 | |
| Deletion | Chr5 | 20434001 | 20561000 | 126999 | 21 |
| Deletion | Chr2 | 21197001 | 21217000 | 19999 | 2 |
| Deletion | Chr1 | 22146001 | 22175000 | 28999 | 4 |
| Deletion | Chr7 | 22691001 | 22713000 | 21999 | 4 |
| Deletion | Chr12 | 23324998 | 23325003 | 5 | |
| Deletion | Chr12 | 24420001 | 24449000 | 28999 | 3 |
| Deletion | Chr7 | 26188965 | 26188978 | 13 | |
| Deletion | Chr12 | 26464107 | 26464121 | 14 | |
| Deletion | Chr11 | 26601834 | 26601835 | 1 | |
| Deletion | Chr2 | 27887001 | 27908000 | 20999 | 4 |
| Deletion | Chr11 | 28996150 | 28996151 | 1 | |
| Deletion | Chr6 | 29376763 | 29376764 | 1 | |
| Deletion | Chr4 | 29649444 | 29649445 | 1 | |
| Deletion | Chr2 | 31417001 | 31481000 | 63999 | 9 |
Insertions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21243030 | 21243032 | 3 | |
| Insertion | Chr1 | 28904030 | 28904033 | 4 | |
| Insertion | Chr10 | 13395688 | 13395691 | 4 | |
| Insertion | Chr10 | 5512865 | 5512865 | 1 | |
| Insertion | Chr11 | 24199989 | 24199990 | 2 | |
| Insertion | Chr11 | 27065088 | 27065088 | 1 | |
| Insertion | Chr12 | 20633789 | 20633797 | 9 | |
| Insertion | Chr12 | 25785111 | 25785130 | 20 | |
| Insertion | Chr3 | 20862130 | 20862130 | 1 | |
| Insertion | Chr3 | 2355512 | 2355600 | 89 | |
| Insertion | Chr3 | 32944306 | 32944306 | 1 | |
| Insertion | Chr4 | 14748003 | 14748004 | 2 | |
| Insertion | Chr4 | 29203953 | 29203953 | 1 | LOC_Os04g48970 |
| Insertion | Chr4 | 35261571 | 35261571 | 1 | |
| Insertion | Chr5 | 25750379 | 25750380 | 2 | |
| Insertion | Chr5 | 9952385 | 9952385 | 1 | |
| Insertion | Chr6 | 12007334 | 12007334 | 1 | |
| Insertion | Chr7 | 11810008 | 11810058 | 51 | |
| Insertion | Chr8 | 27970626 | 27970627 | 2 | |
| Insertion | Chr8 | 6341171 | 6341172 | 2 | |
| Insertion | Chr9 | 22463082 | 22463090 | 9 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 9980951 | 10795252 | |
| Inversion | Chr8 | 25051223 | 25051957 | |
| Inversion | Chr6 | 29889698 | 29890423 | LOC_Os06g49330 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4006323 | Chr4 | 33382101 | LOC_Os12g07900 |
| Translocation | Chr12 | 4006867 | Chr4 | 33382100 | LOC_Os12g07900 |
| Translocation | Chr11 | 10848049 | Chr1 | 17616091 | LOC_Os11g19030 |
| Translocation | Chr11 | 10848486 | Chr1 | 17616073 | |
| Translocation | Chr5 | 14223371 | Chr4 | 50479 | |
| Translocation | Chr11 | 19363264 | Chr2 | 31415402 | |
| Translocation | Chr11 | 19363270 | Chr2 | 31461361 | LOC_Os02g51370 |
| Translocation | Chr11 | 24709249 | Chr7 | 7793849 | |
| Translocation | Chr8 | 28122946 | Chr7 | 840673 | LOC_Os07g02440 |
| Translocation | Chr8 | 28122960 | Chr7 | 840680 | LOC_Os07g02440 |
| Translocation | Chr11 | 28480779 | Chr4 | 3329195 |
