Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2990-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2990-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 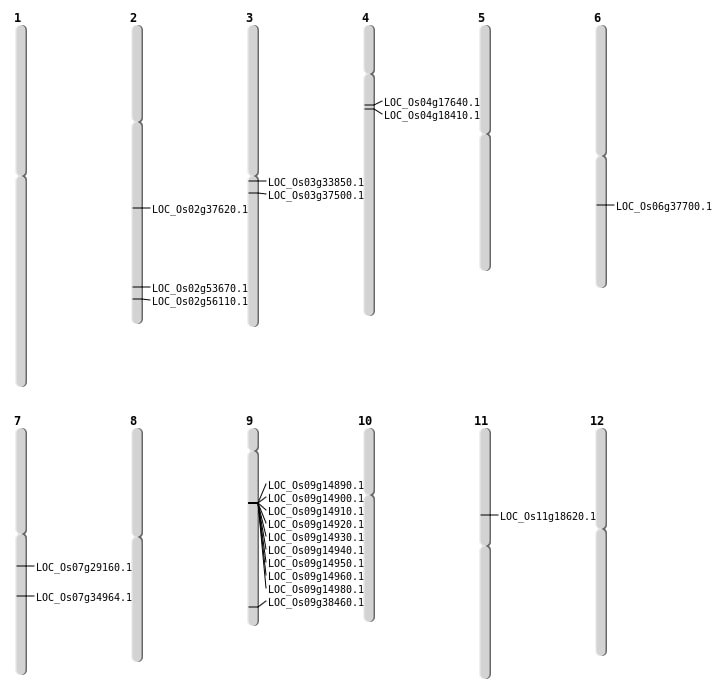
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20220161 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 42242731 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 2027363 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 2577869 | G-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 11769047 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8580138 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8580139 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 19053396 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 31287287 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 31287292 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 32829940 | C-T | HET | STOP_GAINED | LOC_Os02g53670 |
| SBS | Chr3 | 19334859 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33850 |
| SBS | Chr3 | 30231722 | C-A | HOMO | INTRON | |
| SBS | Chr3 | 32312070 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 8306710 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 11161 | G-A | HET | INTRON | |
| SBS | Chr4 | 11276116 | A-G | HOMO | INTRON | |
| SBS | Chr4 | 11315442 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 11762406 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 21001443 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 6051618 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 10423124 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11702945 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 15037276 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 17070887 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g29160 |
| SBS | Chr7 | 20922876 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 12142592 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 22146025 | T-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 20728418 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 20736075 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 20736076 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 22139014 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38460 |
| SBS | Chr9 | 22835000 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4339880 | G-T | HOMO | INTRON |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 133782 | 133783 | 1 | |
| Deletion | Chr4 | 161685 | 161687 | 2 | |
| Deletion | Chr12 | 2269355 | 2269364 | 9 | |
| Deletion | Chr4 | 2453589 | 2453594 | 5 | |
| Deletion | Chr4 | 7631491 | 7631492 | 1 | |
| Deletion | Chr9 | 8877001 | 8926000 | 48999 | 5 |
| Deletion | Chr9 | 8929001 | 8977000 | 47999 | 4 |
| Deletion | Chr4 | 10166401 | 10166404 | 3 | LOC_Os04g18410 |
| Deletion | Chr4 | 10383555 | 10383572 | 17 | |
| Deletion | Chr11 | 10507936 | 10507937 | 1 | LOC_Os11g18620 |
| Deletion | Chr6 | 12471155 | 12471194 | 39 | |
| Deletion | Chr9 | 14504962 | 14504967 | 5 | |
| Deletion | Chr3 | 15590401 | 15590421 | 20 | |
| Deletion | Chr4 | 16011404 | 16011411 | 7 | |
| Deletion | Chr4 | 17141828 | 17141829 | 1 | |
| Deletion | Chr12 | 18036999 | 18037090 | 91 | |
| Deletion | Chr5 | 18386840 | 18386846 | 6 | |
| Deletion | Chr1 | 18491074 | 18491075 | 1 | |
| Deletion | Chr10 | 18704607 | 18704608 | 1 | |
| Deletion | Chr2 | 20052263 | 20052266 | 3 | |
| Deletion | Chr3 | 20810654 | 20810655 | 1 | LOC_Os03g37500 |
| Deletion | Chr2 | 22703382 | 22703418 | 36 | LOC_Os02g37620 |
| Deletion | Chr8 | 23258540 | 23258541 | 1 | |
| Deletion | Chr4 | 24123260 | 24123265 | 5 | |
| Deletion | Chr2 | 31633991 | 31634005 | 14 | |
| Deletion | Chr2 | 32830517 | 32830529 | 12 | LOC_Os02g53670 |
| Deletion | Chr2 | 34343782 | 34343783 | 1 | LOC_Os02g56110 |
| Deletion | Chr1 | 38122405 | 38122410 | 5 |
Insertions: 7
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 9658619 | 9674988 | LOC_Os04g17640 |
| Inversion | Chr7 | 20928560 | 20947499 | LOC_Os07g34964 |
| Inversion | Chr7 | 20928562 | 20947500 | LOC_Os07g34964 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 22337468 | Chr4 | 4257957 | LOC_Os06g37700 |
