Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3033-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3033-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 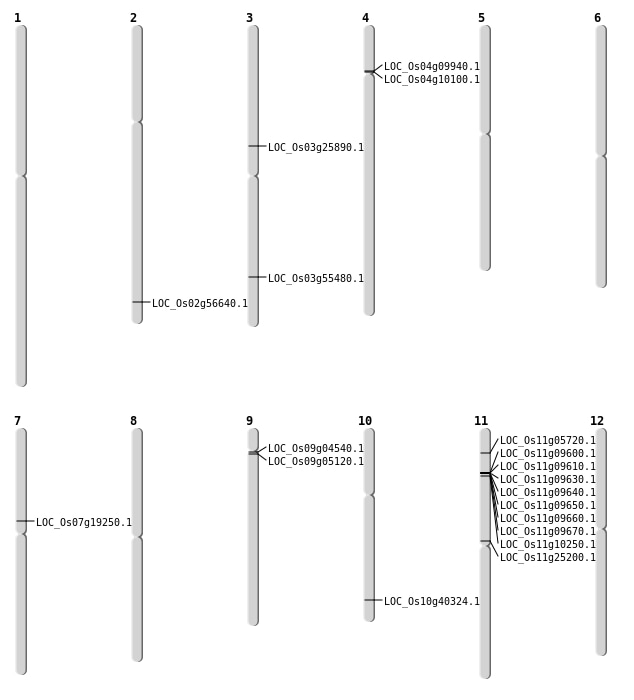
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18288964 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 18621509 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 26418162 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 28198570 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 34537978 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 34537980 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 34537993 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11427416 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2621675 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g05720 |
| SBS | Chr12 | 1990486 | G-A | HET | INTRON | |
| SBS | Chr12 | 2091194 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 2091213 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 21744241 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 15276145 | A-T | HOMO | INTRON | |
| SBS | Chr2 | 15276146 | A-T | HOMO | INTRON | |
| SBS | Chr2 | 7570970 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 17598819 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 17954274 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 30188330 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 6973619 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 8333095 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 24429411 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 12140270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 14801228 | G-C | HOMO | INTRON | |
| SBS | Chr5 | 17212235 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 17212236 | G-A | HOMO | INTRON | |
| SBS | Chr5 | 2841477 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 6779328 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 10587973 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 23837737 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 13439453 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 27746761 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 2208603 | T-C | HET | INTRON | |
| SBS | Chr9 | 2421331 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04540 |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 30726 | 30728 | 2 | |
| Deletion | Chr11 | 904712 | 904728 | 16 | |
| Deletion | Chr3 | 2404430 | 2404432 | 2 | |
| Deletion | Chr9 | 2748231 | 2748246 | 15 | LOC_Os09g05120 |
| Deletion | Chr1 | 4106497 | 4106513 | 16 | |
| Deletion | Chr9 | 4819910 | 4819911 | 1 | |
| Deletion | Chr11 | 5129001 | 5167000 | 37999 | 8 |
| Deletion | Chr4 | 5345561 | 5345573 | 12 | LOC_Os04g09940 |
| Deletion | Chr9 | 5755891 | 5755892 | 1 | |
| Deletion | Chr7 | 6384552 | 6384554 | 2 | |
| Deletion | Chr10 | 6479811 | 6479812 | 1 | |
| Deletion | Chr8 | 6836488 | 6836497 | 9 | |
| Deletion | Chr7 | 7576079 | 7576170 | 91 | |
| Deletion | Chr5 | 7853811 | 7853813 | 2 | |
| Deletion | Chr12 | 9705869 | 9705870 | 1 | |
| Deletion | Chr3 | 11458164 | 11458198 | 34 | |
| Deletion | Chr6 | 12165467 | 12165468 | 1 | |
| Deletion | Chr7 | 12175990 | 12175991 | 1 | |
| Deletion | Chr4 | 14111089 | 14111090 | 1 | |
| Deletion | Chr5 | 14189946 | 14190007 | 61 | |
| Deletion | Chr11 | 14358048 | 14358049 | 1 | LOC_Os11g25200 |
| Deletion | Chr3 | 14809336 | 14809337 | 1 | LOC_Os03g25890 |
| Deletion | Chr2 | 15296576 | 15296579 | 3 | |
| Deletion | Chr11 | 16179152 | 16179236 | 84 | |
| Deletion | Chr11 | 16179161 | 16179187 | 26 | |
| Deletion | Chr3 | 18608704 | 18608712 | 8 | |
| Deletion | Chr4 | 20546191 | 20546192 | 1 | |
| Deletion | Chr4 | 21362561 | 21362569 | 8 | |
| Deletion | Chr12 | 24805609 | 24805610 | 1 | |
| Deletion | Chr8 | 25351287 | 25351292 | 5 | |
| Deletion | Chr3 | 31563484 | 31563485 | 1 | LOC_Os03g55480 |
| Deletion | Chr2 | 33006858 | 33006860 | 2 | |
| Deletion | Chr1 | 34049175 | 34049176 | 1 | |
| Deletion | Chr3 | 34061138 | 34061155 | 17 | |
| Deletion | Chr2 | 34727471 | 34727475 | 4 | LOC_Os02g56640 |
| Deletion | Chr3 | 35328950 | 35329042 | 92 | |
| Deletion | Chr4 | 35493094 | 35493100 | 6 |
Insertions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 18277414 | 18277433 | 20 | |
| Insertion | Chr1 | 31139013 | 31139013 | 1 | |
| Insertion | Chr1 | 36337021 | 36337022 | 2 | |
| Insertion | Chr1 | 39383077 | 39383077 | 1 | |
| Insertion | Chr1 | 8612246 | 8612247 | 2 | |
| Insertion | Chr10 | 11140452 | 11140452 | 1 | |
| Insertion | Chr10 | 22625204 | 22625205 | 2 | |
| Insertion | Chr10 | 23013436 | 23013447 | 12 | |
| Insertion | Chr10 | 6931474 | 6931474 | 1 | |
| Insertion | Chr3 | 22607473 | 22607484 | 12 | |
| Insertion | Chr3 | 30815278 | 30815281 | 4 | |
| Insertion | Chr3 | 31280028 | 31280037 | 10 | |
| Insertion | Chr4 | 17512339 | 17512339 | 1 | |
| Insertion | Chr4 | 5453520 | 5453523 | 4 | LOC_Os04g10100 |
| Insertion | Chr5 | 10152588 | 10152588 | 1 | |
| Insertion | Chr5 | 169904 | 169906 | 3 | |
| Insertion | Chr5 | 7413933 | 7414011 | 79 | |
| Insertion | Chr5 | 9504585 | 9504585 | 1 | |
| Insertion | Chr6 | 8656151 | 8656174 | 24 | |
| Insertion | Chr7 | 11399650 | 11399662 | 13 | LOC_Os07g19250 |
| Insertion | Chr7 | 17617229 | 17617318 | 90 | |
| Insertion | Chr7 | 6072669 | 6072670 | 2 | |
| Insertion | Chr8 | 12575994 | 12575994 | 1 | |
| Insertion | Chr8 | 18205386 | 18205391 | 6 | |
| Insertion | Chr8 | 24834151 | 24834152 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 38551352 | 38553087 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 12598601 | Chr4 | 3152664 | |
| Translocation | Chr6 | 22249449 | Chr4 | 20828281 | |
| Translocation | Chr12 | 27020465 | Chr10 | 21613382 | LOC_Os10g40324 |
