Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3129-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3129-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 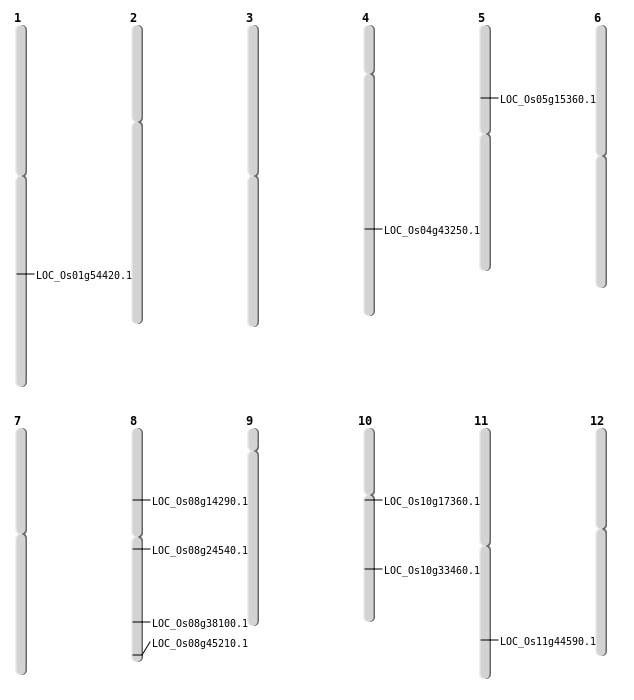
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19171498 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 21673211 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 24563964 | A-G | HET | INTRON | |
| SBS | Chr1 | 30467802 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 31311517 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g54420 |
| SBS | Chr1 | 37051310 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17461092 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 17619515 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g33460 |
| SBS | Chr10 | 17619526 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g33460 |
| SBS | Chr11 | 16654678 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 25383038 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 14915973 | G-A | HET | INTRON | |
| SBS | Chr2 | 17151179 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20282309 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 5808896 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 29962688 | G-A | HET | INTRON | |
| SBS | Chr3 | 337641 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 34345407 | C-T | HET | INTRON | |
| SBS | Chr4 | 1214492 | G-A | HET | INTRON | |
| SBS | Chr4 | 25587586 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43250 |
| SBS | Chr4 | 27821386 | A-C | HET | INTRON | |
| SBS | Chr4 | 32523044 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 16655202 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 17434756 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 17273608 | C-T | HET | INTRON | |
| SBS | Chr6 | 17534062 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20462834 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 6168588 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 20515939 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 28390844 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g45210 |
| SBS | Chr9 | 1567425 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 8046391 | A-C | HOMO | INTRON |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 885040 | 885074 | 34 | |
| Deletion | Chr1 | 1259504 | 1259923 | 419 | |
| Deletion | Chr4 | 2627645 | 2627646 | 1 | |
| Deletion | Chr6 | 4304251 | 4304259 | 8 | |
| Deletion | Chr4 | 4853057 | 4853143 | 86 | |
| Deletion | Chr7 | 7281586 | 7281647 | 61 | |
| Deletion | Chr4 | 8158805 | 8158806 | 1 | |
| Deletion | Chr11 | 8689441 | 8689533 | 92 | |
| Deletion | Chr10 | 8748930 | 8748931 | 1 | LOC_Os10g17360 |
| Deletion | Chr6 | 9599772 | 9599773 | 1 | |
| Deletion | Chr10 | 9660742 | 9660746 | 4 | |
| Deletion | Chr2 | 10331673 | 10331685 | 12 | |
| Deletion | Chr4 | 11945405 | 11945406 | 1 | |
| Deletion | Chr7 | 12154985 | 12154986 | 1 | |
| Deletion | Chr11 | 12533012 | 12533013 | 1 | |
| Deletion | Chr7 | 14189955 | 14189956 | 1 | |
| Deletion | Chr10 | 14797291 | 14797302 | 11 | |
| Deletion | Chr8 | 14820167 | 14820232 | 65 | LOC_Os08g24540 |
| Deletion | Chr5 | 15242374 | 15242375 | 1 | |
| Deletion | Chr8 | 17964425 | 17964451 | 26 | |
| Deletion | Chr4 | 18479384 | 18479385 | 1 | |
| Deletion | Chr11 | 20263500 | 20263506 | 6 | |
| Deletion | Chr5 | 21626190 | 21626191 | 1 | |
| Deletion | Chr7 | 23404504 | 23404505 | 1 | |
| Deletion | Chr11 | 25063526 | 25063576 | 50 | |
| Deletion | Chr12 | 26542753 | 26542754 | 1 | |
| Deletion | Chr8 | 27143959 | 27143961 | 2 | |
| Deletion | Chr5 | 28982946 | 28982948 | 2 | |
| Deletion | Chr2 | 30240549 | 30240550 | 1 | |
| Deletion | Chr1 | 31954509 | 31954512 | 3 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 11844458 | 11844458 | 1 | |
| Insertion | Chr10 | 5412334 | 5412334 | 1 | |
| Insertion | Chr11 | 26147165 | 26147165 | 1 | |
| Insertion | Chr12 | 14518922 | 14518922 | 1 | |
| Insertion | Chr2 | 17551991 | 17551991 | 1 | |
| Insertion | Chr4 | 15266164 | 15266164 | 1 | |
| Insertion | Chr4 | 32912475 | 32912475 | 1 | |
| Insertion | Chr5 | 10007414 | 10007414 | 1 | |
| Insertion | Chr8 | 24138366 | 24138366 | 1 | LOC_Os08g38100 |
| Insertion | Chr8 | 6843935 | 6843935 | 1 | |
| Insertion | Chr9 | 5748486 | 5748486 | 1 |
No Inversion
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3655572 | Chr1 | 35235637 | |
| Translocation | Chr8 | 6761169 | Chr5 | 8422521 | |
| Translocation | Chr8 | 8561243 | Chr1 | 14847279 | LOC_Os08g14290 |
| Translocation | Chr5 | 8695387 | Chr4 | 20317846 | LOC_Os05g15360 |
| Translocation | Chr9 | 9371551 | Chr7 | 4204682 | |
| Translocation | Chr9 | 11379209 | Chr6 | 7774338 | |
| Translocation | Chr11 | 13397516 | Chr2 | 2470221 | |
| Translocation | Chr10 | 22550420 | Chr1 | 2473716 | |
| Translocation | Chr11 | 26960166 | Chr5 | 27709523 | LOC_Os11g44590 |
| Translocation | Chr4 | 33803295 | Chr1 | 34719682 |
