Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3132-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3132-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 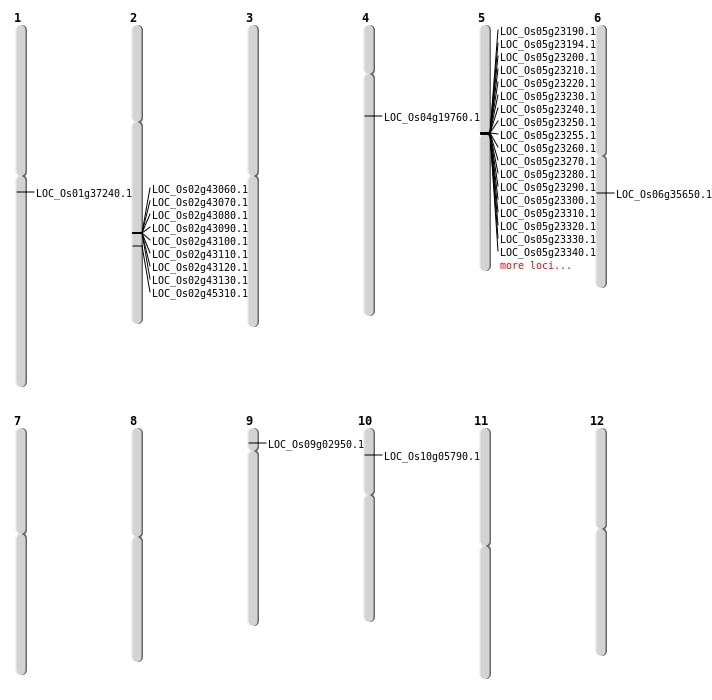
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10360688 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 2527332 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3438530 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 43213370 | G-T | HOMO | INTRON | |
| SBS | Chr10 | 12407415 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14420073 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 15893826 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 17196175 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 2923535 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g05790 |
| SBS | Chr10 | 692051 | C-A | HET | INTRON | |
| SBS | Chr11 | 11179822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 503407 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 636343 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 2882360 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 25311755 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 26392004 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 28264979 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 9762694 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29779159 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 4470467 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 7100797 | T-A | HET | INTRON | |
| SBS | Chr3 | 8489585 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 18989030 | C-A | HOMO | INTRON | |
| SBS | Chr4 | 22286402 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 26216887 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26928713 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 33593427 | T-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 9445501 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 13475397 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 13475398 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 27992134 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 7193104 | G-A | HET | INTRON | |
| SBS | Chr5 | 7940170 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 12656488 | C-G | HET | INTRON | |
| SBS | Chr6 | 23066967 | G-A | HET | INTRON | |
| SBS | Chr7 | 16126345 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18226425 | G-A | HET | INTRON | |
| SBS | Chr7 | 19309344 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr7 | 8921462 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 9890367 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 14169693 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 1894288 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 27733481 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr8 | 4068135 | T-A | HET | INTRON | |
| SBS | Chr8 | 7021900 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 13874133 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 14077814 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 4789703 | C-T | HET | SYNONYMOUS_CODING |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1207891 | 1207896 | 5 | |
| Deletion | Chr12 | 3246954 | 3246963 | 9 | |
| Deletion | Chr6 | 5006634 | 5006641 | 7 | |
| Deletion | Chr2 | 5348110 | 5348112 | 2 | |
| Deletion | Chr7 | 5829124 | 5829125 | 1 | |
| Deletion | Chr3 | 6214489 | 6214491 | 2 | |
| Deletion | Chr5 | 7658697 | 7658703 | 6 | |
| Deletion | Chr3 | 9177569 | 9177570 | 1 | |
| Deletion | Chr4 | 11039949 | 11039963 | 14 | LOC_Os04g19760 |
| Deletion | Chr12 | 11290147 | 11290154 | 7 | |
| Deletion | Chr5 | 13220001 | 13464000 | 243999 | 36 |
| Deletion | Chr7 | 13363615 | 13363616 | 1 | |
| Deletion | Chr5 | 13892651 | 13892656 | 5 | |
| Deletion | Chr10 | 14421597 | 14421611 | 14 | |
| Deletion | Chr7 | 14622473 | 14622474 | 1 | |
| Deletion | Chr2 | 16066376 | 16066378 | 2 | |
| Deletion | Chr7 | 16886672 | 16886673 | 1 | |
| Deletion | Chr9 | 17919458 | 17919464 | 6 | |
| Deletion | Chr8 | 18315316 | 18315317 | 1 | |
| Deletion | Chr12 | 20762965 | 20762966 | 1 | |
| Deletion | Chr6 | 20809362 | 20809364 | 2 | LOC_Os06g35650 |
| Deletion | Chr11 | 21364706 | 21364712 | 6 | |
| Deletion | Chr6 | 24442161 | 24442162 | 1 | |
| Deletion | Chr2 | 25920001 | 25975000 | 54999 | 9 |
| Deletion | Chr2 | 35408079 | 35408089 | 10 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 20797926 | 20797927 | 2 | LOC_Os01g37240 |
| Insertion | Chr2 | 13374696 | 13374696 | 1 | |
| Insertion | Chr2 | 360680 | 360680 | 1 | |
| Insertion | Chr5 | 12748341 | 12748369 | 29 | |
| Insertion | Chr5 | 346092 | 346093 | 2 | |
| Insertion | Chr6 | 9003437 | 9003490 | 54 | |
| Insertion | Chr9 | 13250037 | 13250042 | 6 | |
| Insertion | Chr9 | 1383674 | 1383674 | 1 | LOC_Os09g02950 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 23312769 | 23564908 | |
| Inversion | Chr7 | 23312781 | 23564909 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1001714 | Chr2 | 15292875 | |
| Translocation | Chr7 | 1188610 | Chr4 | 18475332 | |
| Translocation | Chr7 | 5440325 | Chr6 | 7372684 | |
| Translocation | Chr12 | 8348298 | Chr10 | 15364393 |
