Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3165-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3165-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 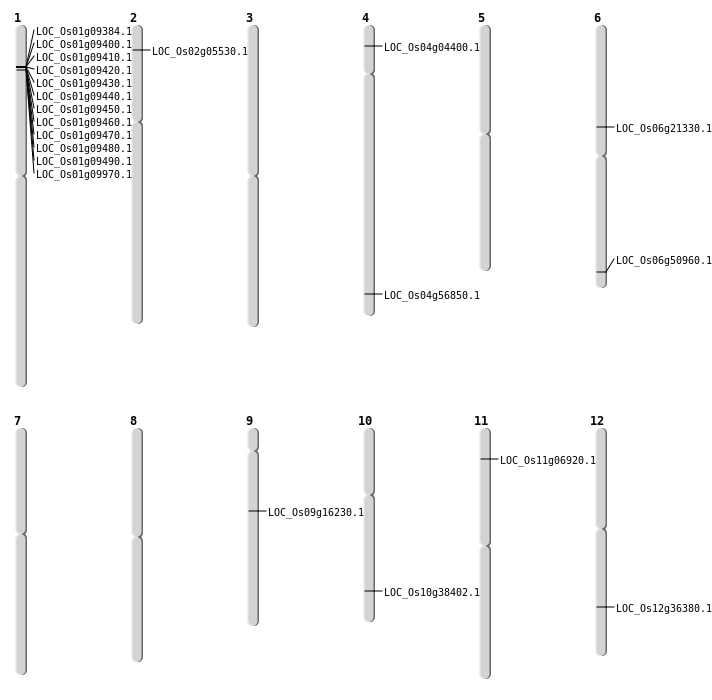
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14628700 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 32519950 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 16055731 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 16795333 | C-T | HET | INTRON | |
| SBS | Chr10 | 16795337 | G-T | HET | INTRON | |
| SBS | Chr10 | 20516655 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 20516656 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g38402 |
| SBS | Chr11 | 10162797 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 12829976 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 11937854 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 18002467 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 22272617 | C-T | HOMO | STOP_GAINED | LOC_Os12g36380 |
| SBS | Chr12 | 4311369 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 7473375 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 14378463 | C-A | HOMO | INTRON | |
| SBS | Chr2 | 16911492 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 19989165 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28083502 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28822167 | A-C | HET | INTRON | |
| SBS | Chr3 | 14149567 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 28200199 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29652658 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 1605980 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 2095308 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04400 |
| SBS | Chr4 | 33889385 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g56850 |
| SBS | Chr4 | 8916912 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 16890867 | T-A | HET | INTRON | |
| SBS | Chr5 | 6519464 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 7855875 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 12203578 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 12318935 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g21330 |
| SBS | Chr6 | 19580338 | A-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 7345165 | G-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 7782338 | G-A | HET | INTRON | |
| SBS | Chr7 | 1436141 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 18984209 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 26701797 | G-C | HET | INTRON | |
| SBS | Chr7 | 27739298 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11216690 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 6569800 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 6622486 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 9909398 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g16230 |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 769349 | 769358 | 9 | |
| Deletion | Chr11 | 3402958 | 3402959 | 1 | LOC_Os11g06920 |
| Deletion | Chr1 | 4767001 | 4777000 | 9999 | LOC_Os01g09384 |
| Deletion | Chr1 | 4781001 | 4842000 | 60999 | 11 |
| Deletion | Chr9 | 7356049 | 7356916 | 867 | |
| Deletion | Chr12 | 7453314 | 7453318 | 4 | |
| Deletion | Chr7 | 8165671 | 8165672 | 1 | |
| Deletion | Chr8 | 8387525 | 8387526 | 1 | |
| Deletion | Chr10 | 9404388 | 9404411 | 23 | |
| Deletion | Chr4 | 11666862 | 11666866 | 4 | |
| Deletion | Chr2 | 11931805 | 11931806 | 1 | |
| Deletion | Chr11 | 12157466 | 12157467 | 1 | |
| Deletion | Chr5 | 13812545 | 13812580 | 35 | |
| Deletion | Chr7 | 13903213 | 13903215 | 2 | |
| Deletion | Chr9 | 15848817 | 15848858 | 41 | |
| Deletion | Chr6 | 16844212 | 16844219 | 7 | |
| Deletion | Chr10 | 20184321 | 20184371 | 50 | |
| Deletion | Chr10 | 20502589 | 20502590 | 1 | |
| Deletion | Chr6 | 20738754 | 20738756 | 2 | |
| Deletion | Chr7 | 21189500 | 21189502 | 2 | |
| Deletion | Chr8 | 22128427 | 22128431 | 4 | |
| Deletion | Chr7 | 24644201 | 24644202 | 1 | |
| Deletion | Chr6 | 24982279 | 24982286 | 7 | |
| Deletion | Chr6 | 25662382 | 25662391 | 9 | |
| Deletion | Chr6 | 25988358 | 25988359 | 1 | |
| Deletion | Chr8 | 26803958 | 26803970 | 12 | |
| Deletion | Chr8 | 28307514 | 28307515 | 1 | |
| Deletion | Chr6 | 28569001 | 28586000 | 16999 | LOC_Os06g50960 |
| Deletion | Chr4 | 28811609 | 28811614 | 5 | |
| Deletion | Chr2 | 30406424 | 30406426 | 2 | |
| Deletion | Chr4 | 32893831 | 32893832 | 1 | |
| Deletion | Chr1 | 35896812 | 35896813 | 1 |
Insertions: 12
No Inversion
Translocations: 13
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 34037 | Chr3 | 16882320 | |
| Translocation | Chr3 | 4463815 | Chr2 | 2674139 | LOC_Os02g05530 |
| Translocation | Chr9 | 5118424 | Chr1 | 26919364 | |
| Translocation | Chr9 | 5118428 | Chr1 | 26927575 | |
| Translocation | Chr11 | 7158246 | Chr10 | 6978185 | |
| Translocation | Chr8 | 10343873 | Chr1 | 11602709 | |
| Translocation | Chr8 | 10388279 | Chr1 | 11602708 | |
| Translocation | Chr11 | 13485820 | Chr2 | 18272186 | |
| Translocation | Chr12 | 15953891 | Chr9 | 1340903 | |
| Translocation | Chr4 | 16579755 | Chr2 | 21668128 | |
| Translocation | Chr7 | 22010603 | Chr3 | 8428627 | |
| Translocation | Chr11 | 22193654 | Chr4 | 5327082 | |
| Translocation | Chr11 | 23694688 | Chr1 | 25246388 |
