Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3171-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3171-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 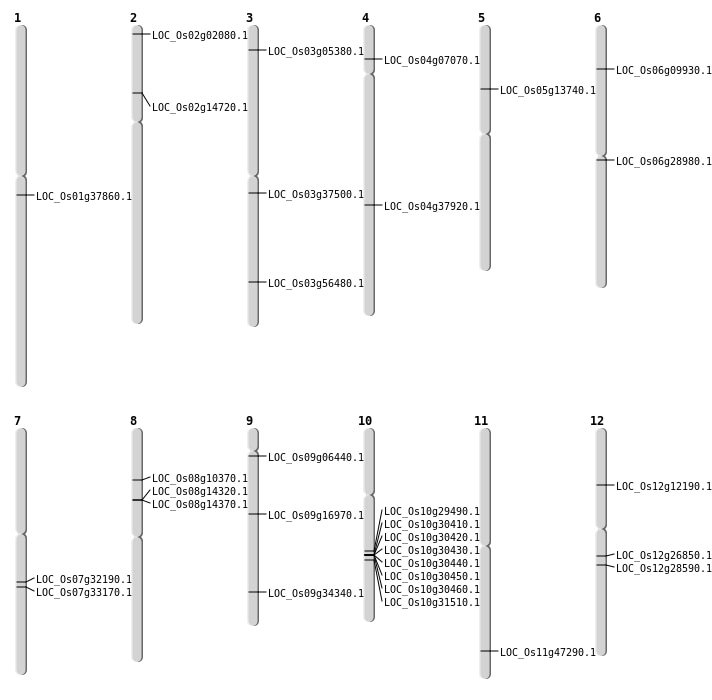
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21190438 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g37860 |
| SBS | Chr1 | 28906157 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32570228 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 8146677 | A-T | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 8857408 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 10752594 | G-A | HET | INTRON | |
| SBS | Chr11 | 17742647 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 2197842 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 5785518 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 9985118 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 23779575 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 5021885 | A-G | HET | INTRON | |
| SBS | Chr12 | 6691932 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g12190 |
| SBS | Chr12 | 6691933 | A-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 28492086 | C-A | HET | INTRON | |
| SBS | Chr2 | 8121376 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g14720 |
| SBS | Chr2 | 9093184 | C-A | HET | INTRON | |
| SBS | Chr3 | 7900689 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 8536373 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 3749247 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g07070 |
| SBS | Chr4 | 9571247 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 1844545 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13710812 | G-A | HET | INTRON | |
| SBS | Chr6 | 16595957 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 20565593 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 31199284 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 5061877 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g09930 |
| SBS | Chr6 | 5413710 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 19135873 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32190 |
| SBS | Chr7 | 24044283 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 9446152 | A-C | HOMO | INTRON | |
| SBS | Chr8 | 9446153 | C-T | HOMO | INTRON |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1505729 | 1505743 | 14 | |
| Deletion | Chr10 | 4566881 | 4566894 | 13 | |
| Deletion | Chr1 | 4961944 | 4961952 | 8 | |
| Deletion | Chr5 | 6082375 | 6082376 | 1 | |
| Deletion | Chr6 | 6273653 | 6273654 | 1 | |
| Deletion | Chr9 | 6312370 | 6312379 | 9 | |
| Deletion | Chr6 | 6390954 | 6390964 | 10 | |
| Deletion | Chr5 | 7615570 | 7615571 | 1 | LOC_Os05g13740 |
| Deletion | Chr6 | 8679783 | 8679808 | 25 | |
| Deletion | Chr2 | 9264620 | 9264621 | 1 | |
| Deletion | Chr7 | 9407154 | 9407155 | 1 | |
| Deletion | Chr4 | 9633809 | 9633824 | 15 | |
| Deletion | Chr9 | 10375433 | 10375474 | 41 | LOC_Os09g16970 |
| Deletion | Chr4 | 11919744 | 11919755 | 11 | |
| Deletion | Chr4 | 12180464 | 12180465 | 1 | |
| Deletion | Chr11 | 12595172 | 12595173 | 1 | |
| Deletion | Chr2 | 13032346 | 13032347 | 1 | |
| Deletion | Chr5 | 13871527 | 13871528 | 1 | |
| Deletion | Chr5 | 13948039 | 13948098 | 59 | |
| Deletion | Chr2 | 14730535 | 14730537 | 2 | |
| Deletion | Chr12 | 15733344 | 15733348 | 4 | LOC_Os12g26850 |
| Deletion | Chr10 | 15809001 | 15846000 | 36999 | 6 |
| Deletion | Chr6 | 16047945 | 16047967 | 22 | |
| Deletion | Chr2 | 16439674 | 16439675 | 1 | |
| Deletion | Chr6 | 16517917 | 16517925 | 8 | LOC_Os06g28980 |
| Deletion | Chr12 | 16910114 | 16910117 | 3 | LOC_Os12g28590 |
| Deletion | Chr2 | 18782842 | 18782879 | 37 | |
| Deletion | Chr3 | 20246870 | 20246872 | 2 | |
| Deletion | Chr9 | 20274111 | 20274114 | 3 | LOC_Os09g34340 |
| Deletion | Chr3 | 20810676 | 20810677 | 1 | LOC_Os03g37500 |
| Deletion | Chr4 | 21212730 | 21212731 | 1 | |
| Deletion | Chr10 | 22408168 | 22408169 | 1 | |
| Deletion | Chr11 | 24200200 | 24200206 | 6 | |
| Deletion | Chr11 | 26705709 | 26705710 | 1 | |
| Deletion | Chr11 | 27065427 | 27065430 | 3 | |
| Deletion | Chr5 | 28885194 | 28885197 | 3 | |
| Deletion | Chr6 | 29639996 | 29640003 | 7 | |
| Deletion | Chr3 | 32187619 | 32187637 | 18 | LOC_Os03g56480 |
Insertions: 8
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2642983 | 3249893 | LOC_Os03g05380 |
| Inversion | Chr3 | 2647456 | 3250004 | |
| Inversion | Chr8 | 8582395 | 8865174 | LOC_Os08g14320 |
| Inversion | Chr8 | 8582399 | 8624298 | 2 |
| Inversion | Chr10 | 15137651 | 15336193 | |
| Inversion | Chr10 | 15315791 | 15336189 | LOC_Os10g29490 |
Translocations: 20
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 4131080 | Chr2 | 24389251 | |
| Translocation | Chr10 | 4528653 | Chr5 | 5203626 | |
| Translocation | Chr9 | 4945340 | Chr7 | 10938238 | |
| Translocation | Chr10 | 5039112 | Chr5 | 15327794 | |
| Translocation | Chr8 | 6079397 | Chr7 | 19805110 | 2 |
| Translocation | Chr5 | 6098903 | Chr4 | 5793780 | |
| Translocation | Chr6 | 7782063 | Chr1 | 32154696 | |
| Translocation | Chr9 | 10898021 | Chr1 | 13547187 | |
| Translocation | Chr10 | 15137662 | Chr3 | 2642998 | LOC_Os03g05380 |
| Translocation | Chr12 | 15684244 | Chr9 | 7416630 | |
| Translocation | Chr10 | 15810800 | Chr2 | 25422835 | LOC_Os10g30410 |
| Translocation | Chr10 | 15846308 | Chr4 | 22418587 | |
| Translocation | Chr10 | 15865228 | Chr4 | 22418575 | |
| Translocation | Chr8 | 15958057 | Chr2 | 610890 | LOC_Os02g02080 |
| Translocation | Chr7 | 16998626 | Chr4 | 22551334 | LOC_Os04g37920 |
| Translocation | Chr10 | 18185378 | Chr9 | 3032276 | LOC_Os09g06440 |
| Translocation | Chr12 | 18397016 | Chr4 | 35416079 | |
| Translocation | Chr8 | 28197584 | Chr3 | 1961324 | |
| Translocation | Chr11 | 28436131 | Chr4 | 22510409 | LOC_Os11g47290 |
| Translocation | Chr2 | 29427397 | Chr1 | 37455144 |
