Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3175-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3175-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 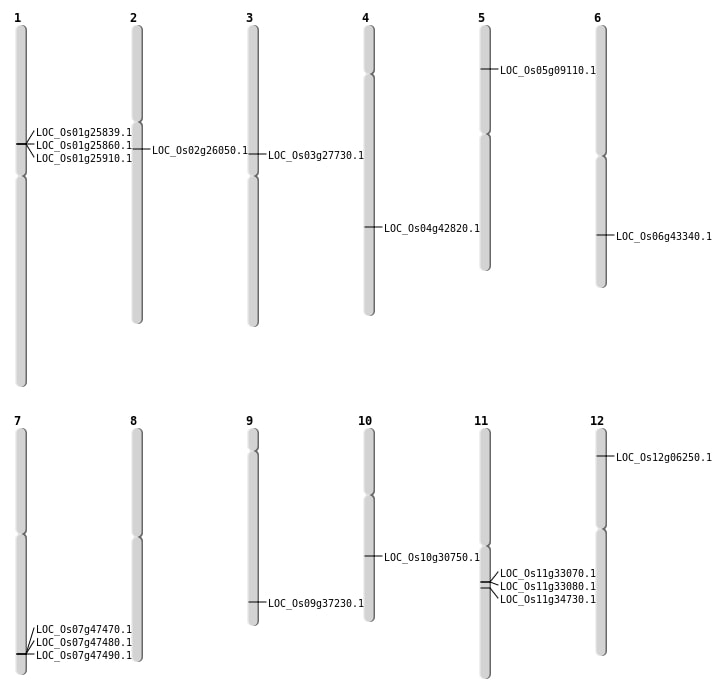
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 41106524 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15804469 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15804470 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 16020973 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30750 |
| SBS | Chr10 | 16020974 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30750 |
| SBS | Chr10 | 4198383 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 9857555 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 26103768 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 30785315 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 31800664 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 33038089 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 34586365 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 20162424 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 25345431 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g42820 |
| SBS | Chr4 | 25919625 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 2654663 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 29372487 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 34604070 | A-T | HET | INTRON | |
| SBS | Chr5 | 17899782 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 5600933 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 7611148 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3468251 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 6959951 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 7075672 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 14088476 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15450152 | T-A | HET | INTRON | |
| SBS | Chr7 | 20396762 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 18808807 | C-T | HET | INTRON | |
| SBS | Chr8 | 449893 | A-T | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 6304858 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 9191549 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21652881 | T-G | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 314092 | 314094 | 2 | |
| Deletion | Chr11 | 1147053 | 1147055 | 2 | |
| Deletion | Chr5 | 5071078 | 5071079 | 1 | LOC_Os05g09110 |
| Deletion | Chr7 | 7459481 | 7459497 | 16 | |
| Deletion | Chr3 | 7465614 | 7465622 | 8 | |
| Deletion | Chr10 | 7667802 | 7667828 | 26 | |
| Deletion | Chr11 | 8069795 | 8069796 | 1 | |
| Deletion | Chr5 | 8551300 | 8551320 | 20 | |
| Deletion | Chr2 | 9161620 | 9161621 | 1 | |
| Deletion | Chr4 | 11233766 | 11233768 | 2 | |
| Deletion | Chr11 | 11461064 | 11461065 | 1 | |
| Deletion | Chr11 | 12161722 | 12161725 | 3 | |
| Deletion | Chr7 | 13475180 | 13475183 | 3 | |
| Deletion | Chr1 | 13972882 | 13972883 | 1 | |
| Deletion | Chr6 | 14097797 | 14097800 | 3 | |
| Deletion | Chr1 | 14624001 | 14677000 | 52999 | 3 |
| Deletion | Chr8 | 15740446 | 15740448 | 2 | |
| Deletion | Chr7 | 17106340 | 17106341 | 1 | |
| Deletion | Chr11 | 19542001 | 19552000 | 9999 | LOC_Os11g33070 |
| Deletion | Chr11 | 19554001 | 19571000 | 16999 | 2 |
| Deletion | Chr7 | 21452040 | 21452042 | 2 | |
| Deletion | Chr3 | 21534014 | 21534015 | 1 | |
| Deletion | Chr12 | 24096243 | 24096250 | 7 | |
| Deletion | Chr6 | 26052825 | 26052827 | 2 | LOC_Os06g43340 |
| Deletion | Chr7 | 28371001 | 28389000 | 17999 | 3 |
| Deletion | Chr3 | 29966319 | 29966320 | 1 | |
| Deletion | Chr2 | 32366699 | 32366705 | 6 |
Insertions: 9
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 26071640 | 26072003 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 687595 | Chr7 | 11905023 | |
| Translocation | Chr10 | 1703886 | Chr3 | 15891627 | LOC_Os03g27730 |
| Translocation | Chr10 | 1703892 | Chr3 | 15892797 | |
| Translocation | Chr8 | 2233762 | Chr5 | 28810789 | |
| Translocation | Chr12 | 2973692 | Chr11 | 2085432 | LOC_Os12g06250 |
| Translocation | Chr7 | 3512181 | Chr1 | 3439728 | |
| Translocation | Chr3 | 3604333 | Chr2 | 16646840 | |
| Translocation | Chr9 | 6397177 | Chr8 | 17892527 | |
| Translocation | Chr7 | 6594584 | Chr4 | 18466064 | |
| Translocation | Chr12 | 8592419 | Chr2 | 15293061 | LOC_Os02g26050 |
| Translocation | Chr8 | 15901663 | Chr1 | 16530859 | |
| Translocation | Chr7 | 17261705 | Chr4 | 8718617 | |
| Translocation | Chr9 | 21492923 | Chr5 | 706733 | LOC_Os09g37230 |
| Translocation | Chr6 | 22658350 | Chr1 | 300013 |
