Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3177-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3177-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 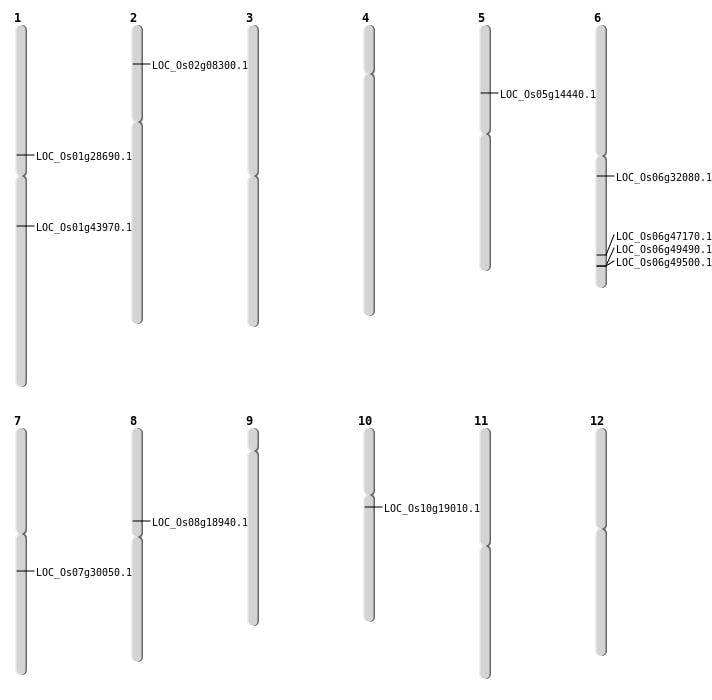
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10837228 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 11096758 | C-T | HET | INTRON | |
| SBS | Chr1 | 14234904 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 26156269 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 26769226 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 9680393 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g19010 |
| SBS | Chr10 | 9680394 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g19010 |
| SBS | Chr11 | 8184966 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 1627890 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 19804462 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 8264642 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 1760887 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 21658126 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 30614363 | G-C | HET | INTRON | |
| SBS | Chr2 | 3746309 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 1189726 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 11921010 | T-A | HET | INTRON | |
| SBS | Chr3 | 14511351 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 15172308 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 24739053 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 25467570 | C-T | HET | INTRON | |
| SBS | Chr3 | 28027433 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 34352986 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 9072259 | C-T | HET | INTRON | |
| SBS | Chr4 | 13190174 | A-T | HET | INTRON | |
| SBS | Chr4 | 4501295 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8681443 | T-A | HET | INTRON | |
| SBS | Chr5 | 8144929 | G-A | HOMO | STOP_GAINED | LOC_Os05g14440 |
| SBS | Chr6 | 12672722 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17652576 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 25302347 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 25302348 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 28604747 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr7 | 10233847 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 17726082 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30050 |
| SBS | Chr7 | 17726083 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30050 |
| SBS | Chr7 | 23081131 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 8437178 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 11298338 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g18940 |
| SBS | Chr8 | 4094558 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 7847293 | C-T | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 167540 | 167543 | 3 | |
| Deletion | Chr1 | 857410 | 857423 | 13 | |
| Deletion | Chr3 | 2347830 | 2347831 | 1 | |
| Deletion | Chr1 | 4208251 | 4208253 | 2 | |
| Deletion | Chr8 | 7348275 | 7348276 | 1 | |
| Deletion | Chr11 | 7731082 | 7731096 | 14 | |
| Deletion | Chr5 | 8164732 | 8164736 | 4 | |
| Deletion | Chr3 | 9069895 | 9069896 | 1 | |
| Deletion | Chr7 | 13313445 | 13313446 | 1 | |
| Deletion | Chr8 | 17044703 | 17044706 | 3 | |
| Deletion | Chr6 | 18655811 | 18655812 | 1 | LOC_Os06g32080 |
| Deletion | Chr4 | 21298816 | 21298822 | 6 | |
| Deletion | Chr8 | 22845090 | 22845091 | 1 | |
| Deletion | Chr3 | 22906322 | 22906368 | 46 | |
| Deletion | Chr4 | 26863945 | 26863946 | 1 | |
| Deletion | Chr11 | 28476996 | 28476997 | 1 | |
| Deletion | Chr6 | 29976001 | 29984000 | 7999 | 3 |
| Deletion | Chr3 | 29988028 | 29988029 | 1 | |
| Deletion | Chr3 | 34340281 | 34340288 | 7 | |
| Deletion | Chr1 | 35894694 | 35894696 | 2 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 25200717 | 25200722 | 6 | LOC_Os01g43970 |
| Insertion | Chr12 | 25289760 | 25289761 | 2 | |
| Insertion | Chr2 | 22745582 | 22745583 | 2 | |
| Insertion | Chr3 | 6607103 | 6607103 | 1 | |
| Insertion | Chr5 | 9790422 | 9790422 | 1 | |
| Insertion | Chr7 | 16239310 | 16239310 | 1 | |
| Insertion | Chr7 | 18898804 | 18898804 | 1 | |
| Insertion | Chr7 | 26511502 | 26511503 | 2 | |
| Insertion | Chr8 | 1226744 | 1226784 | 41 | |
| Insertion | Chr8 | 14632082 | 14632083 | 2 | |
| Insertion | Chr9 | 5959235 | 5959272 | 38 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 4335492 | 4991691 | |
| Inversion | Chr2 | 4412766 | 4990504 | LOC_Os02g08300 |
| Inversion | Chr8 | 13369785 | 13388033 | |
| Inversion | Chr1 | 16066445 | 16133928 | |
| Inversion | Chr6 | 29947123 | 29975225 | |
| Inversion | Chr6 | 29947627 | 29984269 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2806238 | Chr1 | 16065138 | LOC_Os01g28690 |
| Translocation | Chr8 | 2806241 | Chr1 | 16066438 | |
| Translocation | Chr8 | 2806242 | Chr7 | 4108637 | |
| Translocation | Chr7 | 4108810 | Chr1 | 16066437 | |
| Translocation | Chr9 | 8941257 | Chr1 | 30237136 | |
| Translocation | Chr6 | 9896752 | Chr1 | 9951771 | |
| Translocation | Chr11 | 17928209 | Chr3 | 21440143 | |
| Translocation | Chr6 | 25795463 | Chr1 | 16142640 | |
| Translocation | Chr6 | 25795466 | Chr1 | 16140213 |
