Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN318-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN318-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 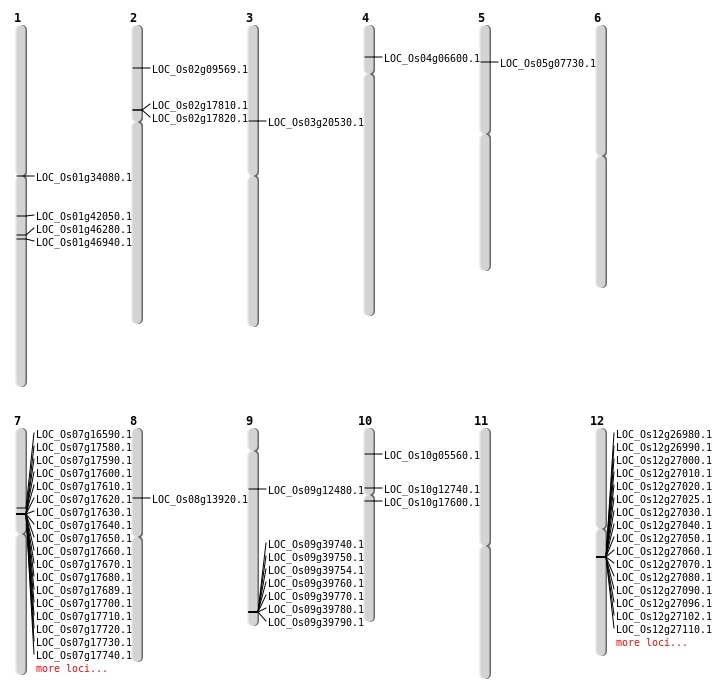
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16821406 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 19069101 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2160883 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 3607089 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 12965851 | G-C | HET | INTRON | |
| SBS | Chr11 | 20091630 | C-A | HET | INTRON | |
| SBS | Chr12 | 17052427 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 2682141 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 12344364 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 26326463 | C-G | HET | INTRON | |
| SBS | Chr3 | 5324649 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr4 | 17277359 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 17429330 | T-G | HOMO | INTRON | |
| SBS | Chr4 | 23925865 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27227829 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 30468888 | A-G | HET | INTRON | |
| SBS | Chr4 | 3480403 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g06600 |
| SBS | Chr4 | 5532590 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 7301071 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 979266 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 14188232 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 4161502 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g07730 |
| SBS | Chr6 | 28486128 | C-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6394367 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 19660493 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 2378126 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 2378127 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 29021001 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 29021002 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 19933281 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 24853587 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 8314448 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 22056638 | T-C | HET | INTRON |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 2720105 | 2720108 | 3 | |
| Deletion | Chr2 | 2808931 | 2808932 | 1 | |
| Deletion | Chr8 | 3445402 | 3445431 | 29 | |
| Deletion | Chr2 | 4679216 | 4679217 | 1 | |
| Deletion | Chr2 | 4927911 | 4927913 | 2 | LOC_Os02g09569 |
| Deletion | Chr3 | 6480310 | 6480311 | 1 | |
| Deletion | Chr3 | 6527757 | 6527758 | 1 | |
| Deletion | Chr10 | 7107343 | 7107348 | 5 | LOC_Os10g12740 |
| Deletion | Chr5 | 7407791 | 7407799 | 8 | |
| Deletion | Chr5 | 7674228 | 7674242 | 14 | |
| Deletion | Chr8 | 8328047 | 8328058 | 11 | LOC_Os08g13920 |
| Deletion | Chr8 | 8649693 | 8649694 | 1 | |
| Deletion | Chr7 | 9730289 | 9730303 | 14 | LOC_Os07g16590 |
| Deletion | Chr2 | 10301344 | 10302882 | 1538 | 2 |
| Deletion | Chr7 | 10379001 | 10564000 | 185000 | 27 |
| Deletion | Chr2 | 11320497 | 11320498 | 1 | |
| Deletion | Chr3 | 11630960 | 11630973 | 13 | LOC_Os03g20530 |
| Deletion | Chr3 | 14203282 | 14203292 | 10 | |
| Deletion | Chr12 | 15059655 | 15059659 | 4 | |
| Deletion | Chr12 | 15804001 | 15966000 | 162000 | 26 |
| Deletion | Chr7 | 16690500 | 16690509 | 9 | LOC_Os07g28510 |
| Deletion | Chr11 | 19039607 | 19039616 | 9 | |
| Deletion | Chr6 | 20794617 | 20794623 | 6 | |
| Deletion | Chr7 | 21206235 | 21206242 | 7 | |
| Deletion | Chr12 | 21526984 | 21526985 | 1 | LOC_Os12g35395 |
| Deletion | Chr9 | 22797335 | 22810198 | 12863 | 6 |
| Deletion | Chr9 | 22812613 | 22813779 | 1166 | LOC_Os09g39790 |
| Deletion | Chr2 | 23905598 | 23905602 | 4 | |
| Deletion | Chr5 | 24899725 | 24899764 | 39 | |
| Deletion | Chr8 | 26028120 | 26028121 | 1 | |
| Deletion | Chr7 | 26285953 | 26285955 | 2 | |
| Deletion | Chr1 | 26791066 | 26791069 | 3 | LOC_Os01g46940 |
| Deletion | Chr4 | 26999151 | 26999158 | 7 | |
| Deletion | Chr4 | 27285748 | 27285803 | 55 | |
| Deletion | Chr3 | 27879587 | 27879594 | 7 | |
| Deletion | Chr4 | 30394797 | 30394800 | 3 | |
| Deletion | Chr3 | 33276672 | 33276674 | 2 | |
| Deletion | Chr1 | 40239284 | 40239285 | 1 | |
| Deletion | Chr1 | 42263783 | 42263786 | 3 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23843460 | 23843460 | 1 | LOC_Os01g42050 |
| Insertion | Chr1 | 8959275 | 8959280 | 6 | |
| Insertion | Chr11 | 17519776 | 17519778 | 3 | |
| Insertion | Chr2 | 9193123 | 9193123 | 1 | |
| Insertion | Chr4 | 23567991 | 23567992 | 2 | |
| Insertion | Chr8 | 16256240 | 16256243 | 4 | |
| Insertion | Chr8 | 3263114 | 3263117 | 4 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 7135477 | 8290316 | LOC_Os09g12480 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2761235 | Chr1 | 26316067 | 2 |
| Translocation | Chr6 | 7209066 | Chr5 | 11443231 | |
| Translocation | Chr10 | 8892685 | Chr7 | 24229874 | 2 |
| Translocation | Chr2 | 10302858 | Chr1 | 18771745 | 2 |
| Translocation | Chr7 | 10539166 | Chr2 | 23019808 | LOC_Os07g17810 |
