Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3184-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3184-S Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 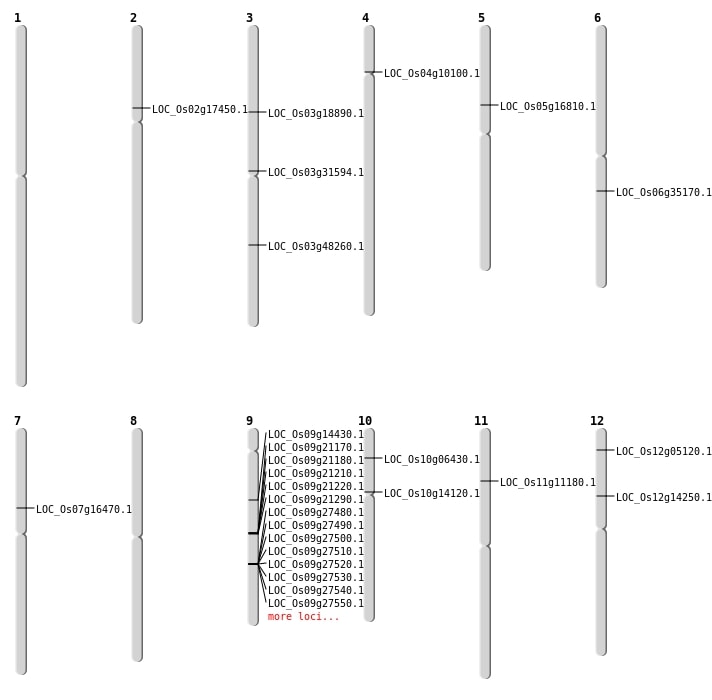
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21523240 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 24287560 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 38815687 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 5632133 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 5827314 | C-G | HOMO | INTRON | |
| SBS | Chr11 | 19194191 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 5800771 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 5800772 | A-C | HOMO | INTRON | |
| SBS | Chr11 | 7649508 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 13484457 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 16521670 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17081738 | C-T | HET | INTRON | |
| SBS | Chr12 | 18739458 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 26848488 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 10040251 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g17450 |
| SBS | Chr2 | 14441144 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 30621811 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 31064371 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr2 | 33473734 | A-T | HET | INTRON | |
| SBS | Chr3 | 10584614 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g18890 |
| SBS | Chr3 | 35895302 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 29301085 | C-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 5452463 | C-A | HOMO | STOP_GAINED | LOC_Os04g10100 |
| SBS | Chr4 | 5452464 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 5452465 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g10100 |
| SBS | Chr4 | 5938724 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 8963848 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 5485524 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 7088705 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 7898925 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 7898926 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 9228110 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 9576344 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g16810 |
| SBS | Chr6 | 19534847 | T-G | HET | INTRON | |
| SBS | Chr7 | 9647207 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16470 |
| SBS | Chr8 | 21087357 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 25773353 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 25854258 | G-A | HET | INTRON | |
| SBS | Chr9 | 14332168 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr9 | 16464619 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 16464624 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 8458486 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 8530264 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g14430 |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1258583 | 1258584 | 1 | |
| Deletion | Chr3 | 1592530 | 1592531 | 1 | |
| Deletion | Chr8 | 1984939 | 1984960 | 21 | |
| Deletion | Chr11 | 2696126 | 2696127 | 1 | |
| Deletion | Chr4 | 2872949 | 2873059 | 110 | |
| Deletion | Chr10 | 3303955 | 3303956 | 1 | LOC_Os10g06430 |
| Deletion | Chr5 | 3921096 | 3921099 | 3 | |
| Deletion | Chr10 | 7566553 | 7566556 | 3 | |
| Deletion | Chr10 | 7665778 | 7665779 | 1 | LOC_Os10g14120 |
| Deletion | Chr12 | 8103151 | 8103153 | 2 | LOC_Os12g14250 |
| Deletion | Chr7 | 8441495 | 8441496 | 1 | |
| Deletion | Chr6 | 8536018 | 8536019 | 1 | |
| Deletion | Chr10 | 8584699 | 8584700 | 1 | |
| Deletion | Chr8 | 11002301 | 11002318 | 17 | |
| Deletion | Chr7 | 11613061 | 11613103 | 42 | |
| Deletion | Chr3 | 12080085 | 12080089 | 4 | |
| Deletion | Chr11 | 12111744 | 12111745 | 1 | |
| Deletion | Chr9 | 12770001 | 12785000 | 14999 | 3 |
| Deletion | Chr9 | 12793001 | 12815000 | 21999 | 2 |
| Deletion | Chr6 | 13770352 | 13770354 | 2 | |
| Deletion | Chr5 | 14281370 | 14281371 | 1 | |
| Deletion | Chr8 | 15571103 | 15571115 | 12 | |
| Deletion | Chr1 | 16317247 | 16317263 | 16 | |
| Deletion | Chr11 | 16553159 | 16553160 | 1 | |
| Deletion | Chr9 | 16684001 | 16871000 | 186999 | 24 |
| Deletion | Chr8 | 16934911 | 16934912 | 1 | |
| Deletion | Chr6 | 17068259 | 17068269 | 10 | |
| Deletion | Chr9 | 17665515 | 17665517 | 2 | |
| Deletion | Chr3 | 18039688 | 18039689 | 1 | LOC_Os03g31594 |
| Deletion | Chr11 | 22525106 | 22525107 | 1 | |
| Deletion | Chr1 | 22549029 | 22549030 | 1 | |
| Deletion | Chr2 | 24448832 | 24448833 | 1 | |
| Deletion | Chr11 | 26602329 | 26602330 | 1 | |
| Deletion | Chr4 | 27952916 | 27952919 | 3 | |
| Deletion | Chr7 | 28838757 | 28838758 | 1 | |
| Deletion | Chr4 | 32798209 | 32798220 | 11 | |
| Deletion | Chr1 | 33576628 | 33576629 | 1 | |
| Deletion | Chr3 | 35617386 | 35617393 | 7 | |
| Deletion | Chr1 | 37466141 | 37466142 | 1 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 2254063 | 2842284 | LOC_Os12g05120 |
| Inversion | Chr12 | 2254070 | 2842289 | LOC_Os12g05120 |
Translocations: 13
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 2487538 | Chr1 | 28210986 | |
| Translocation | Chr8 | 4667316 | Chr5 | 21188254 | |
| Translocation | Chr11 | 5341180 | Chr6 | 8931797 | |
| Translocation | Chr11 | 6190465 | Chr3 | 27458651 | 2 |
| Translocation | Chr4 | 6748884 | Chr2 | 20318706 | |
| Translocation | Chr12 | 12437261 | Chr11 | 19693515 | |
| Translocation | Chr7 | 13781043 | Chr3 | 17297489 | |
| Translocation | Chr3 | 14584527 | Chr2 | 24231589 | |
| Translocation | Chr8 | 15934947 | Chr7 | 6243110 | |
| Translocation | Chr7 | 18371983 | Chr1 | 17686323 | |
| Translocation | Chr11 | 19200649 | Chr9 | 4333976 | |
| Translocation | Chr6 | 20502515 | Chr1 | 11599807 | LOC_Os06g35170 |
| Translocation | Chr6 | 27455075 | Chr1 | 20917541 |
