Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3189-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3189-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 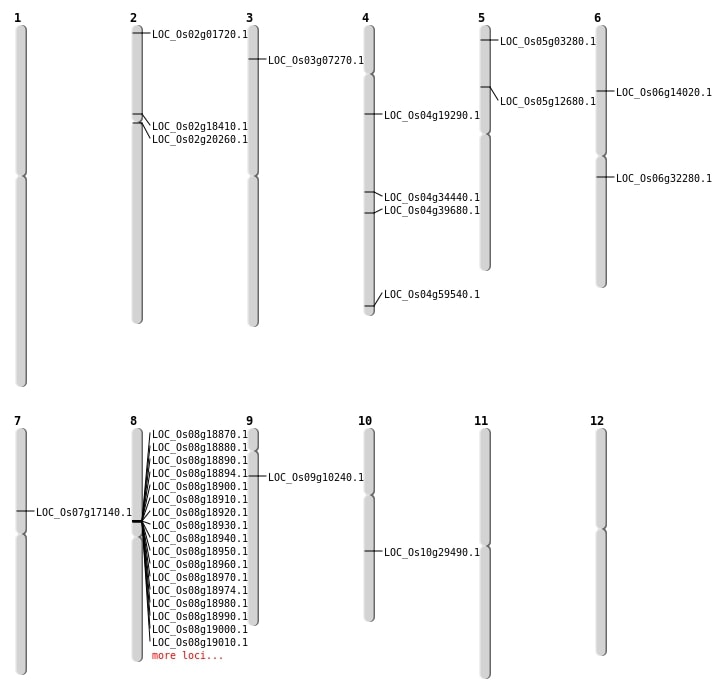
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2979536 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 2979537 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 34116879 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 36174770 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 36174772 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 9205803 | G-A | HET | INTRON | |
| SBS | Chr10 | 12547619 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 15317609 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g29490 |
| SBS | Chr10 | 1902673 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 6068488 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 7431213 | T-C | HET | INTRON | |
| SBS | Chr2 | 10716617 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18410 |
| SBS | Chr2 | 11929320 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g20260 |
| SBS | Chr2 | 17388022 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 25235836 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 25236969 | G-C | HET | INTRON | |
| SBS | Chr2 | 26868785 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 11563539 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 9119815 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 10735771 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19290 |
| SBS | Chr4 | 1407453 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 17264077 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 18720358 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20854182 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g34440 |
| SBS | Chr4 | 33021712 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 35416362 | G-T | HET | STOP_GAINED | LOC_Os04g59540 |
| SBS | Chr4 | 6115387 | C-A | HET | INTRON | |
| SBS | Chr4 | 7013653 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 22821569 | C-G | HET | INTRON | |
| SBS | Chr5 | 25274872 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 11965492 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 19720883 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr6 | 6166128 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 7796111 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g14020 |
| SBS | Chr7 | 14459662 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 14952552 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 26357669 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr7 | 3202351 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 5755553 | T-A | HET | INTRON | |
| SBS | Chr7 | 7508983 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 12349758 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 1299262 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 9916059 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 15389468 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 15393609 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 5578828 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10240 |
| SBS | Chr9 | 6149788 | C-T | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2650176 | 2650342 | 166 | |
| Deletion | Chr5 | 7284220 | 7284222 | 2 | LOC_Os05g12680 |
| Deletion | Chr6 | 7574936 | 7574943 | 7 | |
| Deletion | Chr2 | 7831460 | 7831461 | 1 | |
| Deletion | Chr8 | 7903182 | 7903215 | 33 | |
| Deletion | Chr4 | 9825672 | 9825673 | 1 | |
| Deletion | Chr5 | 9893210 | 9893211 | 1 | |
| Deletion | Chr7 | 9953194 | 9953195 | 1 | |
| Deletion | Chr7 | 10071021 | 10071030 | 9 | LOC_Os07g17140 |
| Deletion | Chr8 | 11254001 | 11371000 | 116999 | 22 |
| Deletion | Chr8 | 17056951 | 17056952 | 1 | LOC_Os08g27990 |
| Deletion | Chr9 | 17819066 | 17819072 | 6 | |
| Deletion | Chr12 | 18408539 | 18408541 | 2 | |
| Deletion | Chr6 | 18792766 | 18792767 | 1 | LOC_Os06g32280 |
| Deletion | Chr4 | 23642474 | 23642483 | 9 | LOC_Os04g39680 |
| Deletion | Chr2 | 25212454 | 25212455 | 1 | |
| Deletion | Chr1 | 30337256 | 30337261 | 5 | |
| Deletion | Chr3 | 31639821 | 31639822 | 1 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 21112606 | 21112659 | 54 | |
| Insertion | Chr11 | 25644115 | 25644186 | 72 | |
| Insertion | Chr2 | 32366711 | 32366712 | 2 | |
| Insertion | Chr4 | 31708405 | 31708427 | 23 | |
| Insertion | Chr4 | 5224277 | 5224306 | 30 | |
| Insertion | Chr4 | 5413691 | 5413692 | 2 | |
| Insertion | Chr5 | 1329961 | 1329961 | 1 | LOC_Os05g03280 |
| Insertion | Chr5 | 8994340 | 8994341 | 2 | |
| Insertion | Chr6 | 575232 | 575233 | 2 | |
| Insertion | Chr7 | 1340872 | 1340872 | 1 | |
| Insertion | Chr8 | 12039186 | 12039187 | 2 | |
| Insertion | Chr8 | 8606012 | 8606012 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 16470778 | 16842047 | |
| Inversion | Chr2 | 16843860 | 17016145 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 3952271 | Chr1 | 2817593 | |
| Translocation | Chr10 | 15776444 | Chr2 | 397595 | LOC_Os02g01720 |
| Translocation | Chr10 | 17232278 | Chr1 | 7861537 | |
| Translocation | Chr11 | 18646936 | Chr2 | 19206812 | |
| Translocation | Chr6 | 23042479 | Chr3 | 3707180 | LOC_Os03g07270 |
