Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3199-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3199-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 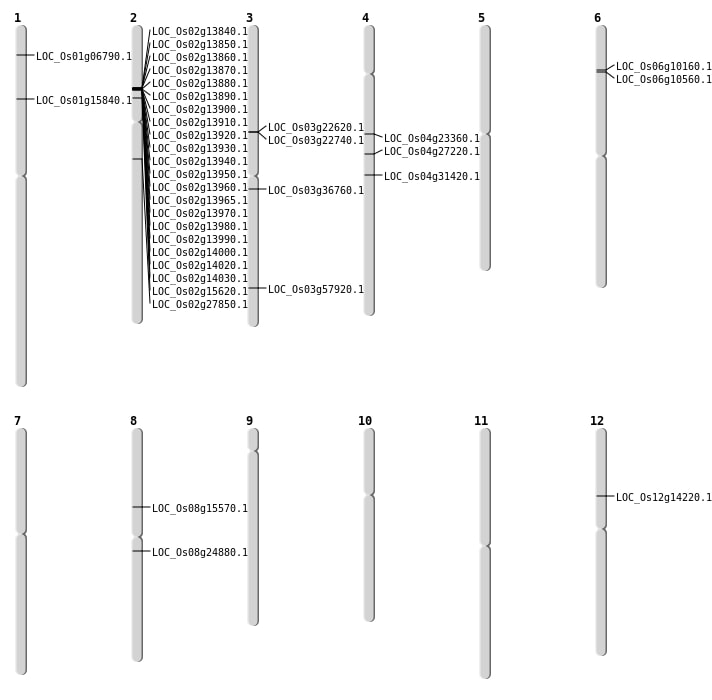
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 36938070 | G-T | HET | INTRON | |
| SBS | Chr1 | 37356987 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 39992773 | C-T | HET | INTRON | |
| SBS | Chr1 | 4097612 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 41159833 | T-C | HET | START_GAINED | |
| SBS | Chr1 | 8924753 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g15840 |
| SBS | Chr10 | 14035188 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 22918128 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 3219097 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 18566870 | G-T | HOMO | INTRON | |
| SBS | Chr12 | 12629589 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 16034199 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 15144643 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 15144646 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 13059010 | A-G | HET | INTRON | |
| SBS | Chr3 | 20394766 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g36760 |
| SBS | Chr3 | 23123639 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 26291189 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 28043010 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 29218789 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 34395775 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 18792746 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g31420 |
| SBS | Chr4 | 19595363 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 28646618 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 32468023 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34215469 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 22502562 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 24990364 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 5719999 | A-T | HET | INTRON | |
| SBS | Chr5 | 9965887 | A-G | HOMO | INTRON | |
| SBS | Chr6 | 15246048 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 5443829 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 29575198 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12324413 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 26742083 | T-A | HET | INTRON | |
| SBS | Chr8 | 27337795 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 5393980 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 9469715 | G-T | HET | STOP_GAINED | LOC_Os08g15570 |
| SBS | Chr9 | 10455630 | T-C | HOMO | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 5198917 | 5198920 | 3 | LOC_Os06g10160 |
| Deletion | Chr6 | 5718500 | 5718502 | 2 | |
| Deletion | Chr2 | 7481001 | 7668000 | 186999 | 21 |
| Deletion | Chr8 | 11002301 | 11002318 | 17 | |
| Deletion | Chr2 | 11120345 | 11120346 | 1 | |
| Deletion | Chr9 | 11462680 | 11462697 | 17 | |
| Deletion | Chr6 | 12194643 | 12194644 | 1 | |
| Deletion | Chr3 | 13134759 | 13134764 | 5 | LOC_Os03g22740 |
| Deletion | Chr4 | 13347826 | 13347831 | 5 | LOC_Os04g23360 |
| Deletion | Chr8 | 15064310 | 15064349 | 39 | LOC_Os08g24880 |
| Deletion | Chr4 | 16088435 | 16088442 | 7 | LOC_Os04g27220 |
| Deletion | Chr2 | 16492894 | 16492897 | 3 | LOC_Os02g27850 |
| Deletion | Chr6 | 17097676 | 17097679 | 3 | |
| Deletion | Chr6 | 17432717 | 17432719 | 2 | |
| Deletion | Chr1 | 18022928 | 18022939 | 11 | |
| Deletion | Chr7 | 18603541 | 18603542 | 1 | |
| Deletion | Chr6 | 18629143 | 18629144 | 1 | |
| Deletion | Chr4 | 24832135 | 24832137 | 2 | |
| Deletion | Chr3 | 26958938 | 26958945 | 7 | |
| Deletion | Chr7 | 28647365 | 28647393 | 28 | |
| Deletion | Chr2 | 31401056 | 31401058 | 2 | |
| Deletion | Chr3 | 32987936 | 32987938 | 2 | LOC_Os03g57920 |
| Deletion | Chr1 | 34224021 | 34224023 | 2 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 32946444 | 32946444 | 1 | |
| Insertion | Chr12 | 8076483 | 8076555 | 73 | LOC_Os12g14220 |
| Insertion | Chr2 | 26485506 | 26485520 | 15 | |
| Insertion | Chr3 | 11157155 | 11157186 | 32 | |
| Insertion | Chr4 | 6856181 | 6856181 | 1 | |
| Insertion | Chr6 | 19777890 | 19777890 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 13037299 | 13061195 | |
| Inversion | Chr3 | 13039678 | 13061215 | LOC_Os03g22620 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 5449824 | Chr3 | 24338834 | LOC_Os06g10560 |
| Translocation | Chr6 | 5451362 | Chr3 | 24338829 | LOC_Os06g10560 |
| Translocation | Chr12 | 8640153 | Chr4 | 2526652 | |
| Translocation | Chr8 | 13361582 | Chr3 | 20015247 | |
| Translocation | Chr8 | 13361586 | Chr3 | 20013315 | |
| Translocation | Chr11 | 27858617 | Chr1 | 3226163 | LOC_Os01g06790 |
