Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3202-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3202-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 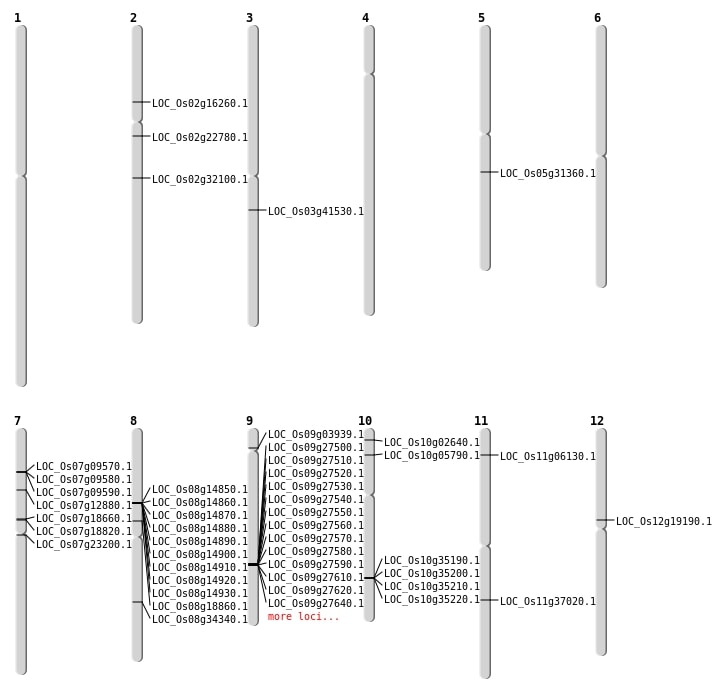
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26237238 | A-T | HET | INTRON | |
| SBS | Chr1 | 39447446 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 41773949 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 18798121 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g35190 |
| SBS | Chr10 | 2922064 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g05790 |
| SBS | Chr11 | 10556833 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21854507 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g37020 |
| SBS | Chr12 | 26067241 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 7132017 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr2 | 13565376 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g22780 |
| SBS | Chr2 | 18944270 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 21010276 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 2427553 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 9382376 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 15818852 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 15818853 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 19773713 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5776600 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5776605 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5776613 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 14372793 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 1568604 | C-T | HET | INTRON | |
| SBS | Chr4 | 17513624 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3516912 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 3516913 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9226723 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 20426915 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 4841108 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 9196706 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 7661209 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18526783 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 25697836 | T-C | HET | INTRON | |
| SBS | Chr8 | 18645766 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 24519321 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 26516176 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 4668835 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 13996825 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 21664146 | T-A | HET | INTRON | |
| SBS | Chr9 | 9316996 | G-A | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1021920 | 1021921 | 1 | LOC_Os10g02640 |
| Deletion | Chr9 | 2007884 | 2007887 | 3 | LOC_Os09g03939 |
| Deletion | Chr6 | 4568925 | 4568948 | 23 | |
| Deletion | Chr8 | 4669311 | 4669320 | 9 | |
| Deletion | Chr9 | 4984052 | 4984058 | 6 | |
| Deletion | Chr7 | 5056001 | 5093000 | 36999 | 4 |
| Deletion | Chr11 | 5087741 | 5087789 | 48 | |
| Deletion | Chr9 | 7607268 | 7607269 | 1 | |
| Deletion | Chr11 | 8438800 | 8438802 | 2 | |
| Deletion | Chr8 | 8927001 | 8998000 | 70999 | 10 |
| Deletion | Chr6 | 9408103 | 9408106 | 3 | |
| Deletion | Chr8 | 10761998 | 10762000 | 2 | |
| Deletion | Chr7 | 11015001 | 11030000 | 14999 | 2 |
| Deletion | Chr12 | 11143037 | 11143044 | 7 | LOC_Os12g19190 |
| Deletion | Chr2 | 13567123 | 13567132 | 9 | |
| Deletion | Chr12 | 16443370 | 16443371 | 1 | |
| Deletion | Chr9 | 16701001 | 16797000 | 95999 | 13 |
| Deletion | Chr9 | 16739566 | 16739567 | 1 | |
| Deletion | Chr9 | 16801001 | 17106000 | 304999 | 54 |
| Deletion | Chr5 | 18228919 | 18228928 | 9 | LOC_Os05g31360 |
| Deletion | Chr10 | 18795001 | 18821000 | 25999 | 4 |
| Deletion | Chr3 | 19722377 | 19722390 | 13 | |
| Deletion | Chr8 | 21557800 | 21557803 | 3 | LOC_Os08g34340 |
| Deletion | Chr1 | 21760214 | 21760224 | 10 | |
| Deletion | Chr5 | 21863696 | 21863697 | 1 | |
| Deletion | Chr3 | 23095183 | 23095186 | 3 | LOC_Os03g41530 |
| Deletion | Chr7 | 26170120 | 26170121 | 1 | |
| Deletion | Chr5 | 29941679 | 29941680 | 1 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 14045450 | 14045461 | 12 | |
| Insertion | Chr12 | 25059689 | 25059710 | 22 | |
| Insertion | Chr2 | 12949264 | 12949265 | 2 | |
| Insertion | Chr2 | 18822027 | 18822027 | 1 | |
| Insertion | Chr2 | 18969464 | 18969466 | 3 | LOC_Os02g32100 |
| Insertion | Chr3 | 25527866 | 25527893 | 28 | |
| Insertion | Chr7 | 17560179 | 17560180 | 2 | |
| Insertion | Chr9 | 11354282 | 11354295 | 14 | |
| Insertion | Chr9 | 19095800 | 19095815 | 16 | |
| Insertion | Chr9 | 9768729 | 9768734 | 6 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 5056154 | 5256982 | |
| Inversion | Chr12 | 6924707 | 7141360 | |
| Inversion | Chr12 | 6924756 | 7141365 | |
| Inversion | Chr2 | 9240368 | 9240597 | LOC_Os02g16260 |
| Inversion | Chr2 | 26544654 | 26544785 | |
| Inversion | Chr2 | 29987904 | 29988184 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 5093027 | Chr6 | 28122203 | |
| Translocation | Chr8 | 8926577 | Chr6 | 25354027 | |
| Translocation | Chr8 | 8998124 | Chr6 | 25353075 | |
| Translocation | Chr7 | 13076809 | Chr6 | 28120968 | LOC_Os07g23200 |
| Translocation | Chr11 | 19200649 | Chr9 | 4333986 |
