Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3211-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3211-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 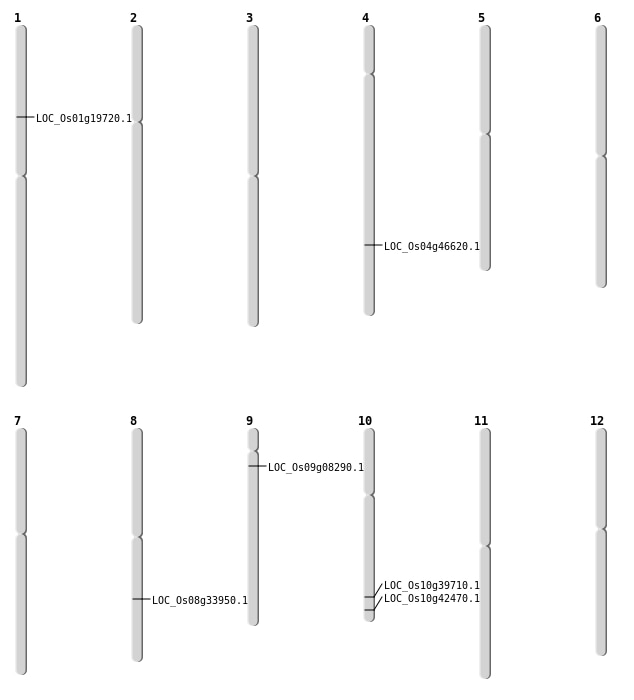
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12032286 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 13378877 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 1814490 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 19537963 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 29542577 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34919998 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 7545707 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 9208525 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 21226056 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g39710 |
| SBS | Chr10 | 22899129 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g42470 |
| SBS | Chr10 | 3230086 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 5085353 | C-T | HET | INTRON | |
| SBS | Chr11 | 1221348 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 3150608 | C-T | HET | INTRON | |
| SBS | Chr12 | 12303348 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 20787002 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 20839151 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 21080041 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 21120915 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 21120923 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 343635 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 3603510 | T-G | HOMO | INTRON | |
| SBS | Chr2 | 10176695 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10827294 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 12690831 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 23513813 | G-A | HET | INTRON | |
| SBS | Chr2 | 29470724 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 4140992 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 6579241 | T-C | HET | INTRON | |
| SBS | Chr3 | 28501288 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 7331711 | A-T | HOMO | INTRON | |
| SBS | Chr4 | 21705714 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 3036608 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 35206730 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 7909466 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 7962905 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 14281927 | G-T | HOMO | INTRON | |
| SBS | Chr5 | 19288487 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 4156021 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 5679153 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8462 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 2102491 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5008414 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 5612175 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr9 | 13141541 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr9 | 4107808 | C-G | HOMO | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1118016 | 1118018 | 2 | |
| Deletion | Chr3 | 2087842 | 2087845 | 3 | |
| Deletion | Chr9 | 4286915 | 4286919 | 4 | LOC_Os09g08290 |
| Deletion | Chr12 | 5029953 | 5029983 | 30 | |
| Deletion | Chr2 | 7832011 | 7832129 | 118 | |
| Deletion | Chr12 | 8437178 | 8437180 | 2 | |
| Deletion | Chr6 | 8600695 | 8600762 | 67 | |
| Deletion | Chr10 | 9013664 | 9013665 | 1 | |
| Deletion | Chr5 | 10194262 | 10194273 | 11 | |
| Deletion | Chr1 | 11184213 | 11184217 | 4 | LOC_Os01g19720 |
| Deletion | Chr6 | 11475343 | 11475349 | 6 | |
| Deletion | Chr8 | 15395839 | 15395841 | 2 | |
| Deletion | Chr3 | 17431215 | 17431216 | 1 | |
| Deletion | Chr5 | 17609575 | 17609577 | 2 | |
| Deletion | Chr11 | 18909432 | 18909434 | 2 | |
| Deletion | Chr10 | 19136053 | 19136054 | 1 | |
| Deletion | Chr5 | 20318970 | 20318975 | 5 | |
| Deletion | Chr2 | 20395414 | 20395416 | 2 | |
| Deletion | Chr2 | 22091028 | 22091029 | 1 | |
| Deletion | Chr12 | 24855745 | 24855746 | 1 | |
| Deletion | Chr4 | 26231761 | 26231763 | 2 | |
| Deletion | Chr11 | 26299305 | 26299306 | 1 | |
| Deletion | Chr12 | 26475409 | 26475410 | 1 | |
| Deletion | Chr6 | 26763371 | 26763375 | 4 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr6 | 27972297 | 27972311 | 14 | |
| Deletion | Chr4 | 28976013 | 28976015 | 2 | |
| Deletion | Chr1 | 38090641 | 38090642 | 1 |
Insertions: 8
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 20788653 | 21122323 | |
| Inversion | Chr12 | 20788665 | 21122332 | |
| Inversion | Chr8 | 21257930 | 21500741 | LOC_Os08g33950 |
| Inversion | Chr8 | 21257946 | 21470779 | LOC_Os08g33950 |
| Inversion | Chr4 | 27641403 | 27641618 | LOC_Os04g46620 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 9573937 | Chr1 | 6540105 | |
| Translocation | Chr12 | 9917951 | Chr4 | 6963432 |
