Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3212-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3212-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 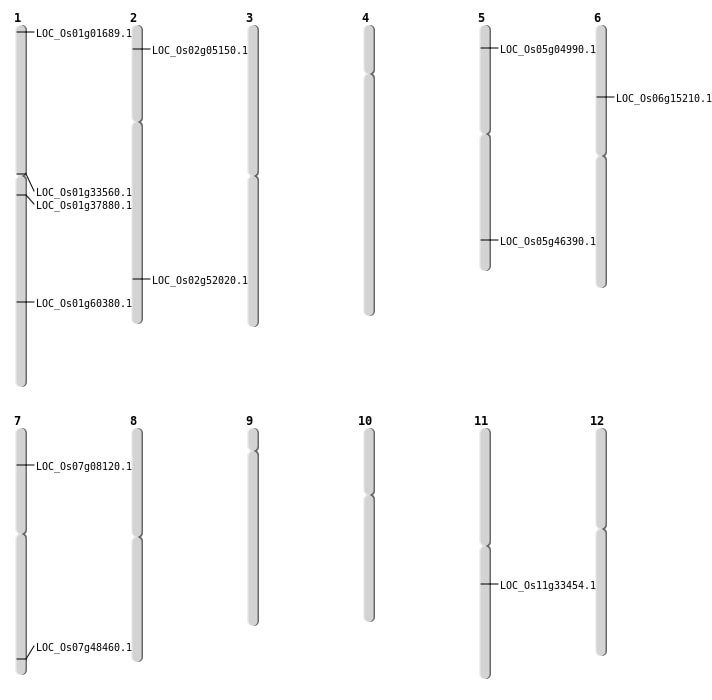
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11165094 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 18472411 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g33560 |
| SBS | Chr1 | 21209598 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g37880 |
| SBS | Chr1 | 338243 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g01689 |
| SBS | Chr1 | 41640734 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 4540825 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 17338261 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3053177 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 7338360 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 17106903 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8116795 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 10723105 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 13733859 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 2463858 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g05150 |
| SBS | Chr2 | 465177 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 16189539 | A-C | HET | INTRON | |
| SBS | Chr3 | 16887670 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 23033924 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 22390494 | A-T | HET | INTRON | |
| SBS | Chr4 | 26533653 | A-C | HET | UTR_5_PRIME | |
| SBS | Chr4 | 6396701 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 10309490 | C-T | HET | INTRON | |
| SBS | Chr5 | 12150645 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20399563 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 2417863 | C-T | HET | STOP_GAINED | LOC_Os05g04990 |
| SBS | Chr5 | 9878616 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13357467 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 24421377 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 27298457 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 27298458 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 4121150 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g08120 |
| SBS | Chr8 | 16090852 | C-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 23639259 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 24339629 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 12443981 | A-T | HOMO | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 461059 | 461061 | 2 | |
| Deletion | Chr5 | 2698006 | 2698008 | 2 | |
| Deletion | Chr11 | 9785812 | 9785824 | 12 | |
| Deletion | Chr4 | 12059341 | 12059342 | 1 | |
| Deletion | Chr11 | 12647428 | 12647429 | 1 | |
| Deletion | Chr7 | 13616580 | 13616581 | 1 | |
| Deletion | Chr10 | 15633338 | 15633340 | 2 | |
| Deletion | Chr10 | 16514062 | 16514068 | 6 | |
| Deletion | Chr8 | 17400550 | 17400555 | 5 | |
| Deletion | Chr2 | 18782842 | 18782879 | 37 | |
| Deletion | Chr11 | 19795127 | 19795130 | 3 | LOC_Os11g33454 |
| Deletion | Chr9 | 20433560 | 20433561 | 1 | |
| Deletion | Chr3 | 22865538 | 22865539 | 1 | |
| Deletion | Chr3 | 23270994 | 23270996 | 2 | |
| Deletion | Chr6 | 25764375 | 25764376 | 1 | |
| Deletion | Chr3 | 26387211 | 26387224 | 13 | |
| Deletion | Chr2 | 31336745 | 31336780 | 35 | |
| Deletion | Chr2 | 31846649 | 31846664 | 15 | LOC_Os02g52020 |
| Deletion | Chr2 | 32425218 | 32425219 | 1 | |
| Deletion | Chr2 | 34541001 | 34541002 | 1 | |
| Deletion | Chr1 | 34925376 | 34925379 | 3 | LOC_Os01g60380 |
| Deletion | Chr1 | 38571602 | 38571620 | 18 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16890676 | 16890676 | 1 | |
| Insertion | Chr10 | 186169 | 186170 | 2 | |
| Insertion | Chr10 | 2770857 | 2770857 | 1 | |
| Insertion | Chr10 | 8231803 | 8231803 | 1 | |
| Insertion | Chr12 | 15679839 | 15679839 | 1 | |
| Insertion | Chr12 | 16866528 | 16866531 | 4 | |
| Insertion | Chr12 | 24971710 | 24971710 | 1 | |
| Insertion | Chr2 | 23336959 | 23336959 | 1 | |
| Insertion | Chr3 | 21360570 | 21360595 | 26 | |
| Insertion | Chr6 | 8619321 | 8619350 | 30 | LOC_Os06g15210 |
| Insertion | Chr8 | 6514991 | 6514991 | 1 | |
| Insertion | Chr8 | 7530426 | 7530426 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 13507809 | 13558994 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1867356 | Chr2 | 16127259 | |
| Translocation | Chr9 | 5979547 | Chr2 | 18788610 | |
| Translocation | Chr7 | 6846435 | Chr1 | 27664994 | |
| Translocation | Chr8 | 15633999 | Chr4 | 3615666 | |
| Translocation | Chr11 | 19842339 | Chr3 | 29848934 | |
| Translocation | Chr10 | 21683380 | Chr5 | 26889343 | LOC_Os05g46390 |
| Translocation | Chr10 | 21683405 | Chr5 | 26849359 | |
| Translocation | Chr7 | 28994733 | Chr4 | 21354356 | LOC_Os07g48460 |
