Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3214-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3214-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 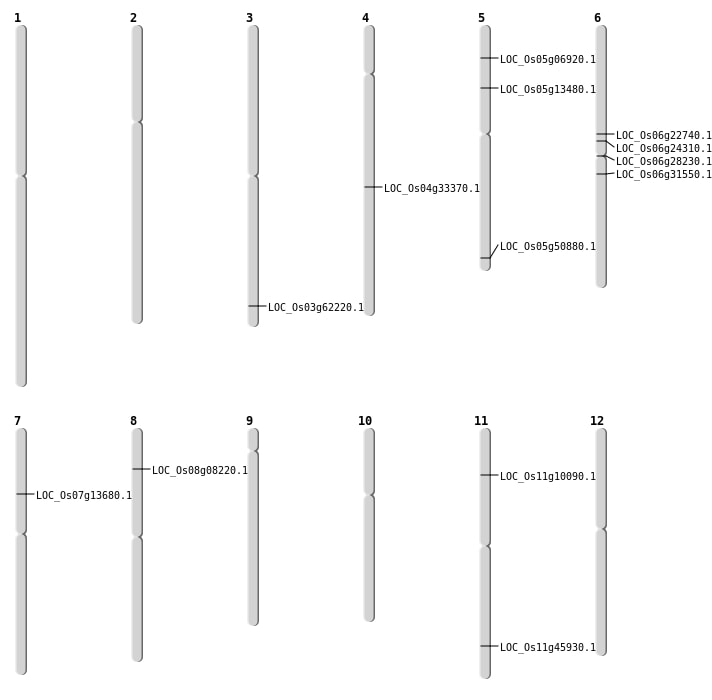
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16511097 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 12427593 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 21020806 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 21692241 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1414535 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 1028752 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 2054745 | A-G | HET | INTRON | |
| SBS | Chr2 | 27088884 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 3945946 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 9133922 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 13345642 | T-A | HET | INTRON | |
| SBS | Chr3 | 13816384 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 17021421 | A-G | HET | INTRON | |
| SBS | Chr3 | 18622778 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 29339449 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 3480573 | A-C | HOMO | INTRON | |
| SBS | Chr4 | 5590542 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 9880631 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 20106525 | G-T | HOMO | INTRON | |
| SBS | Chr5 | 20106528 | A-T | HOMO | INTRON | |
| SBS | Chr5 | 29181221 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g50880 |
| SBS | Chr5 | 29647884 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 3627603 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g06920 |
| SBS | Chr5 | 7476907 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g13480 |
| SBS | Chr6 | 13229449 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g22740 |
| SBS | Chr6 | 13575171 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 16044596 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 26238325 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 6535711 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7853027 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13680 |
| SBS | Chr8 | 12050349 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 3319887 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 9071655 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr8 | 9090038 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 9611570 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5706651 | C-A | HOMO | INTRON |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2650176 | 2650342 | 166 | |
| Deletion | Chr8 | 2861195 | 2861197 | 2 | |
| Deletion | Chr4 | 3210039 | 3210063 | 24 | |
| Deletion | Chr12 | 3898622 | 3898624 | 2 | |
| Deletion | Chr8 | 5415383 | 5415435 | 52 | |
| Deletion | Chr11 | 5447910 | 5447911 | 1 | LOC_Os11g10090 |
| Deletion | Chr7 | 7012063 | 7012064 | 1 | |
| Deletion | Chr6 | 10337836 | 10337837 | 1 | |
| Deletion | Chr4 | 10818290 | 10818291 | 1 | |
| Deletion | Chr11 | 12150216 | 12150247 | 31 | |
| Deletion | Chr8 | 12158178 | 12158179 | 1 | |
| Deletion | Chr4 | 13754369 | 13754370 | 1 | |
| Deletion | Chr4 | 14111089 | 14111090 | 1 | |
| Deletion | Chr4 | 15166736 | 15166738 | 2 | |
| Deletion | Chr6 | 16039524 | 16039533 | 9 | LOC_Os06g28230 |
| Deletion | Chr8 | 17554699 | 17554700 | 1 | |
| Deletion | Chr12 | 17823013 | 17823014 | 1 | |
| Deletion | Chr7 | 21549231 | 21549233 | 2 | |
| Deletion | Chr6 | 22855099 | 22855100 | 1 | |
| Deletion | Chr3 | 22906322 | 22906342 | 20 | |
| Deletion | Chr5 | 23041563 | 23041572 | 9 | |
| Deletion | Chr7 | 23466292 | 23466293 | 1 | |
| Deletion | Chr6 | 26711992 | 26711993 | 1 | |
| Deletion | Chr1 | 27463040 | 27463041 | 1 | |
| Deletion | Chr1 | 28807960 | 28807969 | 9 | |
| Deletion | Chr4 | 33370205 | 33370212 | 7 | |
| Deletion | Chr3 | 33522470 | 33522486 | 16 | |
| Deletion | Chr1 | 34345889 | 34345891 | 2 | |
| Deletion | Chr1 | 36615159 | 36615161 | 2 | |
| Deletion | Chr1 | 42385063 | 42385065 | 2 |
Insertions: 15
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 4703392 | 5070577 | LOC_Os08g08220 |
| Inversion | Chr8 | 4703395 | 5070580 | LOC_Os08g08220 |
| Inversion | Chr6 | 14224981 | 14226227 | LOC_Os06g24310 |
| Inversion | Chr6 | 14224996 | 14226230 | LOC_Os06g24310 |
Translocations: 12
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2610998 | Chr1 | 16147165 | |
| Translocation | Chr11 | 7536128 | Chr6 | 4131691 | |
| Translocation | Chr9 | 8487771 | Chr6 | 18348286 | LOC_Os06g31550 |
| Translocation | Chr10 | 12388291 | Chr1 | 18490297 | |
| Translocation | Chr2 | 12921373 | Chr1 | 33032305 | |
| Translocation | Chr6 | 15501698 | Chr1 | 16931496 | |
| Translocation | Chr6 | 16073303 | Chr5 | 1070147 | |
| Translocation | Chr4 | 20213225 | Chr1 | 11799477 | LOC_Os04g33370 |
| Translocation | Chr7 | 23011906 | Chr4 | 31785700 | |
| Translocation | Chr11 | 27311127 | Chr1 | 20757377 | |
| Translocation | Chr11 | 27794146 | Chr5 | 23586766 | LOC_Os11g45930 |
| Translocation | Chr3 | 35243884 | Chr1 | 35138887 | LOC_Os03g62220 |
