Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3216-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3216-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 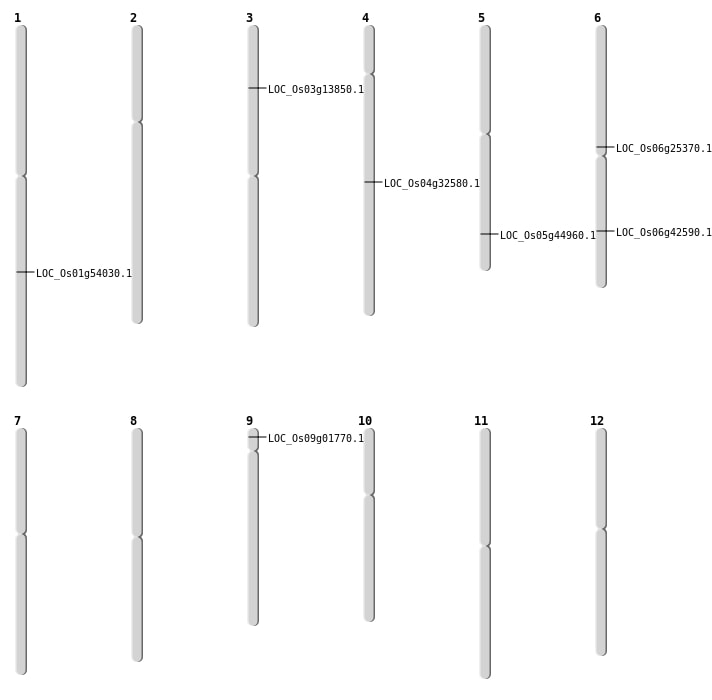
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31077258 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54030 |
| SBS | Chr1 | 31851704 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 13593872 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 10499530 | G-A | HET | INTRON | |
| SBS | Chr12 | 23857679 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 27356501 | C-A | HET | INTRON | |
| SBS | Chr2 | 27811414 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 17277458 | T-G | HET | INTRON | |
| SBS | Chr3 | 19967843 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 6915234 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 28689865 | C-T | HET | INTRON | |
| SBS | Chr4 | 5782414 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6651100 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 17779721 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 24974964 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 26137898 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44960 |
| SBS | Chr5 | 6063009 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 6639963 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 9260889 | A-G | HET | INTRON | |
| SBS | Chr6 | 10742831 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 14860784 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25370 |
| SBS | Chr6 | 15012670 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 17676341 | A-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr6 | 18092647 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 25615439 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42590 |
| SBS | Chr6 | 26366932 | T-C | HET | INTRON | |
| SBS | Chr6 | 28823963 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8058358 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 11746773 | G-A | HET | INTRON | |
| SBS | Chr7 | 21070866 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2756893 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 13609356 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 17273940 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 23134917 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 23407499 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 13830370 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 20281658 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 4378223 | C-T | HOMO | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 2872949 | 2873059 | 110 | |
| Deletion | Chr7 | 4027222 | 4027225 | 3 | |
| Deletion | Chr4 | 4335343 | 4335344 | 1 | |
| Deletion | Chr9 | 4615970 | 4615972 | 2 | |
| Deletion | Chr9 | 5675870 | 5675878 | 8 | |
| Deletion | Chr11 | 7679405 | 7679416 | 11 | |
| Deletion | Chr8 | 10338113 | 10338114 | 1 | |
| Deletion | Chr1 | 12283322 | 12283323 | 1 | |
| Deletion | Chr7 | 13436038 | 13436039 | 1 | |
| Deletion | Chr11 | 15924884 | 15924888 | 4 | |
| Deletion | Chr12 | 16674017 | 16674018 | 1 | |
| Deletion | Chr11 | 17228528 | 17228536 | 8 | |
| Deletion | Chr1 | 17473098 | 17473099 | 1 | |
| Deletion | Chr11 | 18915498 | 18915503 | 5 | |
| Deletion | Chr11 | 19578565 | 19578569 | 4 | |
| Deletion | Chr4 | 19610517 | 19610523 | 6 | LOC_Os04g32580 |
| Deletion | Chr10 | 20845131 | 20845138 | 7 | |
| Deletion | Chr12 | 20997867 | 20997891 | 24 | |
| Deletion | Chr2 | 22517040 | 22517041 | 1 | |
| Deletion | Chr5 | 23813358 | 23813397 | 39 | |
| Deletion | Chr7 | 24028455 | 24028456 | 1 | |
| Deletion | Chr3 | 30627609 | 30627617 | 8 | |
| Deletion | Chr3 | 34562692 | 34562694 | 2 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 5943189 | 5943191 | 3 | |
| Insertion | Chr10 | 87041 | 87138 | 98 | |
| Insertion | Chr11 | 12351117 | 12351120 | 4 | |
| Insertion | Chr12 | 17878258 | 17878258 | 1 | |
| Insertion | Chr12 | 19928397 | 19928397 | 1 | |
| Insertion | Chr12 | 6601194 | 6601194 | 1 | |
| Insertion | Chr12 | 6696620 | 6696623 | 4 | |
| Insertion | Chr2 | 4891348 | 4891349 | 2 | |
| Insertion | Chr4 | 8684439 | 8684439 | 1 | |
| Insertion | Chr5 | 11146517 | 11146519 | 3 | |
| Insertion | Chr6 | 17884405 | 17884405 | 1 | |
| Insertion | Chr6 | 23487453 | 23487493 | 41 | |
| Insertion | Chr6 | 9953444 | 9953444 | 1 | |
| Insertion | Chr8 | 16858425 | 16858520 | 96 | |
| Insertion | Chr8 | 25824153 | 25824154 | 2 | |
| Insertion | Chr9 | 534523 | 534523 | 1 | LOC_Os09g01770 |
No Inversion
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 6237197 | Chr3 | 2806726 | |
| Translocation | Chr5 | 10722802 | Chr4 | 26530139 | |
| Translocation | Chr7 | 11637981 | Chr4 | 9582342 | |
| Translocation | Chr11 | 13498036 | Chr1 | 9838081 | |
| Translocation | Chr5 | 20582313 | Chr3 | 7512118 | LOC_Os03g13850 |
| Translocation | Chr12 | 22022157 | Chr4 | 15489069 | |
| Translocation | Chr6 | 22208177 | Chr4 | 14979951 |
