Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3217-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3217-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 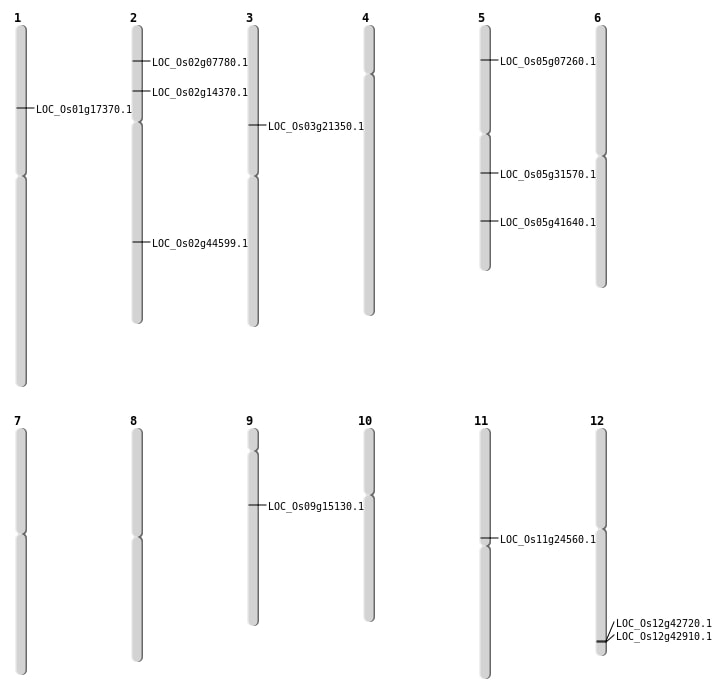
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23752177 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 37885725 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 3905569 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 8104656 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 9999817 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g17370 |
| SBS | Chr10 | 11432976 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15896899 | C-T | HET | INTRON | |
| SBS | Chr10 | 6983727 | T-G | HET | INTRON | |
| SBS | Chr10 | 7258478 | A-G | HET | INTRON | |
| SBS | Chr11 | 14020203 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g24560 |
| SBS | Chr11 | 6404746 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 8645212 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 26543827 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42720 |
| SBS | Chr12 | 26669953 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42910 |
| SBS | Chr2 | 29819616 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 30693157 | C-T | HET | INTRON | |
| SBS | Chr2 | 31647530 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 4382654 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7882705 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14370 |
| SBS | Chr3 | 12211175 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g21350 |
| SBS | Chr3 | 14159388 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 17028548 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 17052951 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 24835372 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 35116275 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5718479 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 5718480 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 5718481 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 9830125 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1230336 | G-T | HET | INTRON | |
| SBS | Chr4 | 482275 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 24380692 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g41640 |
| SBS | Chr6 | 20641101 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21372188 | G-A | HET | INTRON | |
| SBS | Chr7 | 6218672 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 7977557 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 16529890 | C-A | HET | INTRON | |
| SBS | Chr8 | 26773666 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 458364 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9182551 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15130 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 3930731 | 3930742 | 11 | |
| Deletion | Chr2 | 4074300 | 4074301 | 1 | LOC_Os02g07780 |
| Deletion | Chr2 | 4588851 | 4588852 | 1 | |
| Deletion | Chr6 | 4825115 | 4825144 | 29 | |
| Deletion | Chr1 | 5527444 | 5527445 | 1 | |
| Deletion | Chr9 | 7125885 | 7125886 | 1 | |
| Deletion | Chr8 | 10016076 | 10016077 | 1 | |
| Deletion | Chr12 | 10664031 | 10664034 | 3 | |
| Deletion | Chr10 | 11909160 | 11909173 | 13 | |
| Deletion | Chr3 | 12911001 | 12921000 | 9999 | |
| Deletion | Chr5 | 18380812 | 18380813 | 1 | LOC_Os05g31570 |
| Deletion | Chr12 | 23358603 | 23358607 | 4 | |
| Deletion | Chr5 | 24379609 | 24379610 | 1 | LOC_Os05g41640 |
| Deletion | Chr3 | 24587894 | 24587897 | 3 | |
| Deletion | Chr12 | 26843549 | 26843552 | 3 | |
| Deletion | Chr6 | 27749138 | 27749151 | 13 | |
| Deletion | Chr7 | 28388077 | 28388108 | 31 | |
| Deletion | Chr1 | 29540473 | 29540475 | 2 | |
| Deletion | Chr7 | 29678207 | 29678217 | 10 | |
| Deletion | Chr3 | 32781032 | 32781040 | 8 | |
| Deletion | Chr4 | 33125496 | 33125499 | 3 |
Insertions: 11
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 13189569 | 13460356 | |
| Inversion | Chr1 | 13189571 | 13460362 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 769708 | Chr3 | 11386189 | |
| Translocation | Chr10 | 2434259 | Chr4 | 34198565 | |
| Translocation | Chr5 | 3827519 | Chr3 | 12921164 | |
| Translocation | Chr5 | 3841811 | Chr3 | 12910926 | LOC_Os05g07260 |
| Translocation | Chr7 | 10511793 | Chr1 | 19823229 | |
| Translocation | Chr8 | 12580177 | Chr7 | 27346312 | |
| Translocation | Chr10 | 16638143 | Chr2 | 27025135 | LOC_Os02g44599 |
| Translocation | Chr11 | 19735829 | Chr9 | 17966312 |
