Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3219-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3219-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 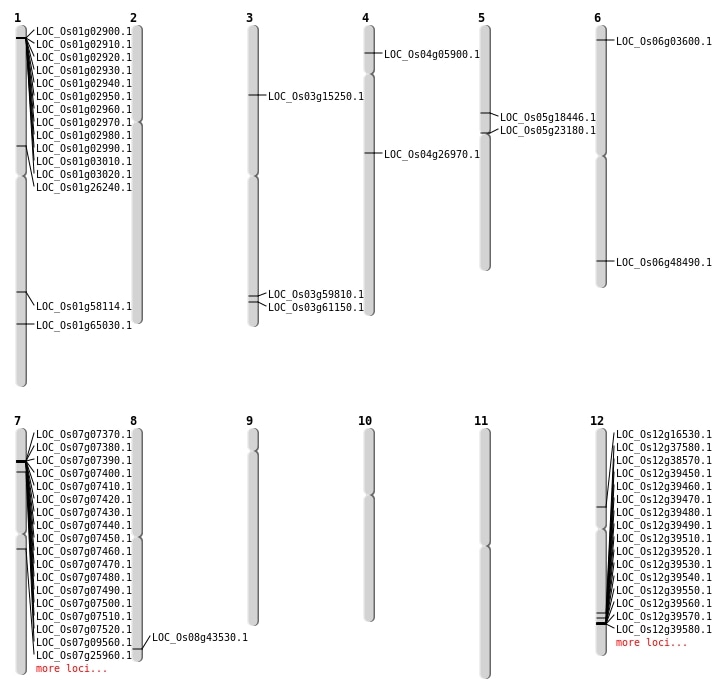
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16268661 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 16716751 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr1 | 3608290 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 37747254 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g65030 |
| SBS | Chr1 | 3951099 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 7976644 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 18941317 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr11 | 13479303 | T-C | HET | INTRON | |
| SBS | Chr11 | 24819626 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1898014 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 23024144 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 4424841 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10946078 | T-A | HET | INTRON | |
| SBS | Chr2 | 15681313 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 34305465 | C-T | HET | INTRON | |
| SBS | Chr2 | 35512140 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 25901518 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 28622379 | G-A | HET | INTRON | |
| SBS | Chr3 | 32364484 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 8340131 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15250 |
| SBS | Chr3 | 8343466 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15250 |
| SBS | Chr4 | 15949793 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g26970 |
| SBS | Chr4 | 29714098 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 31581821 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 3646858 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 25076536 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 5969576 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 5971003 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 2617627 | C-T | HET | INTRON | |
| SBS | Chr7 | 14900021 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25960 |
| SBS | Chr7 | 14900022 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 14900023 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25960 |
| SBS | Chr7 | 19127594 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 19457103 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 21286042 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 28093270 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 28723643 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 6485052 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 13246755 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 15896008 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 1595504 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 20721746 | A-G | HET | INTRON | |
| SBS | Chr9 | 10769552 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17798062 | G-A | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1041001 | 1153000 | 111999 | 13 |
| Deletion | Chr4 | 1472629 | 1472630 | 1 | |
| Deletion | Chr7 | 3683001 | 3750000 | 66999 | 17 |
| Deletion | Chr3 | 4984424 | 4984425 | 1 | |
| Deletion | Chr8 | 6264536 | 6264551 | 15 | |
| Deletion | Chr12 | 8316001 | 8326000 | 9999 | LOC_Os12g16530 |
| Deletion | Chr6 | 9180358 | 9180364 | 6 | |
| Deletion | Chr11 | 9626681 | 9626687 | 6 | |
| Deletion | Chr2 | 10049419 | 10049421 | 2 | |
| Deletion | Chr5 | 10649001 | 10670000 | 20999 | 2 |
| Deletion | Chr5 | 11562686 | 11562717 | 31 | |
| Deletion | Chr4 | 13572220 | 13572221 | 1 | |
| Deletion | Chr3 | 17360896 | 17360919 | 23 | |
| Deletion | Chr7 | 18932001 | 19084000 | 151999 | 23 |
| Deletion | Chr8 | 19282080 | 19282081 | 1 | |
| Deletion | Chr12 | 24273001 | 24511000 | 237999 | 25 |
| Deletion | Chr12 | 24404957 | 24404958 | 1 | |
| Deletion | Chr4 | 25237781 | 25237782 | 1 | |
| Deletion | Chr11 | 25723965 | 25723966 | 1 | |
| Deletion | Chr1 | 33614187 | 33614193 | 6 | LOC_Os01g58114 |
| Deletion | Chr1 | 41394541 | 41394542 | 1 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 15621383 | 15621384 | 2 | |
| Insertion | Chr10 | 17998445 | 17998462 | 18 | |
| Insertion | Chr10 | 912536 | 912536 | 1 | |
| Insertion | Chr11 | 18703647 | 18703648 | 2 | |
| Insertion | Chr5 | 2145586 | 2145591 | 6 | |
| Insertion | Chr5 | 25436725 | 25436725 | 1 | |
| Insertion | Chr6 | 29336028 | 29336065 | 38 | LOC_Os06g48490 |
| Insertion | Chr8 | 12104566 | 12104585 | 20 | |
| Insertion | Chr9 | 987222 | 987233 | 12 |
Inversions: 11
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1429766 | 1429911 | |
| Inversion | Chr11 | 7774137 | 7774334 | |
| Inversion | Chr4 | 14066032 | 14066179 | |
| Inversion | Chr8 | 14369782 | 14370043 | |
| Inversion | Chr8 | 16831953 | 16838329 | |
| Inversion | Chr8 | 16831956 | 16838358 | |
| Inversion | Chr12 | 23073068 | 23073237 | LOC_Os12g37580 |
| Inversion | Chr12 | 23688061 | 23688349 | LOC_Os12g38570 |
| Inversion | Chr1 | 30700285 | 31015377 | |
| Inversion | Chr1 | 30700298 | 31015381 | |
| Inversion | Chr3 | 34733801 | 34734136 | LOC_Os03g61150 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 3055859 | Chr3 | 34057198 | 2 |
| Translocation | Chr12 | 8325719 | Chr6 | 1390531 | LOC_Os06g03600 |
| Translocation | Chr12 | 8327164 | Chr6 | 1390516 | LOC_Os06g03600 |
| Translocation | Chr10 | 8620594 | Chr1 | 12471949 | |
| Translocation | Chr7 | 24086512 | Chr6 | 22803769 | |
| Translocation | Chr12 | 24393553 | Chr6 | 19207692 | |
| Translocation | Chr8 | 27530336 | Chr1 | 23901721 | LOC_Os08g43530 |
