Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3221-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3221-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 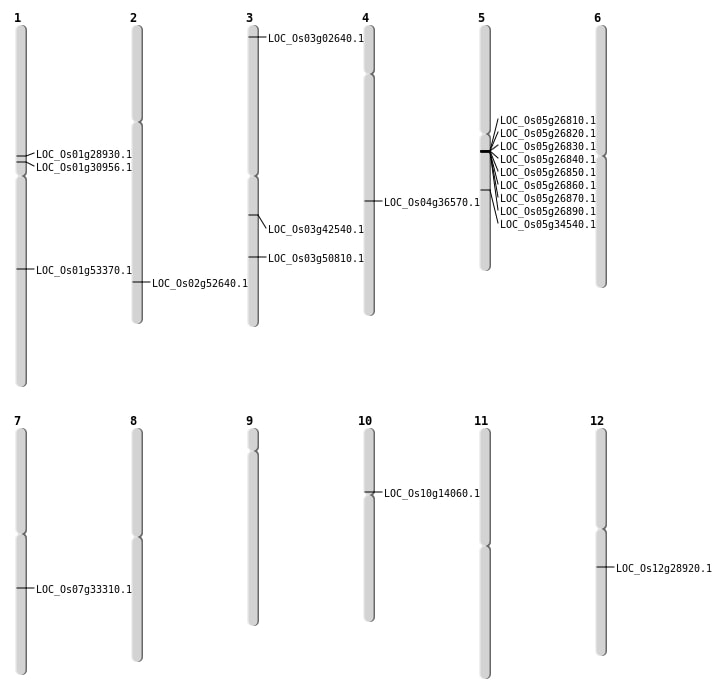
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16905075 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g30956 |
| SBS | Chr1 | 26224148 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 30043430 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 33984316 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 41282843 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 13127226 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 13693169 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 14255434 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 1493385 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7651429 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g14060 |
| SBS | Chr10 | 9639907 | T-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 24871543 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 10987764 | G-T | HET | INTRON | |
| SBS | Chr12 | 11505919 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15827519 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 17093428 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28920 |
| SBS | Chr12 | 20180525 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 11145763 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14621758 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 23673071 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g42540 |
| SBS | Chr3 | 25517655 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 25577455 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 29874538 | G-C | HET | INTRON | |
| SBS | Chr3 | 29902177 | A-C | HET | INTRON | |
| SBS | Chr3 | 3059637 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 35160076 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 5665987 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 10714755 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 31009145 | G-A | HET | INTRON | |
| SBS | Chr4 | 32427590 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 7271076 | A-T | HET | INTRON | |
| SBS | Chr5 | 13040392 | T-C | HET | INTRON | |
| SBS | Chr5 | 25245731 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 25525719 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 25835397 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 11956148 | T-G | HET | INTRON | |
| SBS | Chr6 | 22192620 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 29165227 | T-G | HOMO | INTRON | |
| SBS | Chr6 | 3077381 | G-C | HET | INTRON | |
| SBS | Chr7 | 23025543 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2514793 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27276651 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 9247934 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 10852320 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 18783316 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 18783317 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 2316747 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 4140038 | A-T | HET | INTRON | |
| SBS | Chr8 | 4140041 | T-C | HET | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 926274 | 926291 | 17 | |
| Deletion | Chr7 | 2099830 | 2099834 | 4 | |
| Deletion | Chr7 | 6082689 | 6082781 | 92 | |
| Deletion | Chr11 | 7700797 | 7700799 | 2 | |
| Deletion | Chr4 | 9493146 | 9493148 | 2 | |
| Deletion | Chr2 | 9610500 | 9610506 | 6 | |
| Deletion | Chr6 | 12006873 | 12006889 | 16 | |
| Deletion | Chr8 | 13818626 | 13818633 | 7 | |
| Deletion | Chr4 | 14891750 | 14891751 | 1 | |
| Deletion | Chr5 | 15552001 | 15615000 | 62999 | 8 |
| Deletion | Chr4 | 16204951 | 16204956 | 5 | |
| Deletion | Chr1 | 18477918 | 18477936 | 18 | |
| Deletion | Chr2 | 20743416 | 20743417 | 1 | |
| Deletion | Chr7 | 22603298 | 22603299 | 1 | |
| Deletion | Chr4 | 24853582 | 24853597 | 15 | |
| Deletion | Chr5 | 25466917 | 25466918 | 1 | |
| Deletion | Chr7 | 27759503 | 27759504 | 1 | |
| Deletion | Chr3 | 29018335 | 29018338 | 3 | LOC_Os03g50810 |
| Deletion | Chr1 | 30668094 | 30668106 | 12 | LOC_Os01g53370 |
| Deletion | Chr2 | 32191001 | 32198000 | 6999 | 2 |
| Deletion | Chr4 | 34447462 | 34447463 | 1 | |
| Deletion | Chr1 | 40653094 | 40653102 | 8 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 17096932 | 17096932 | 1 | LOC_Os12g28920 |
| Insertion | Chr2 | 2106504 | 2106505 | 2 | |
| Insertion | Chr2 | 27827307 | 27827350 | 44 | |
| Insertion | Chr3 | 36205284 | 36205286 | 3 | |
| Insertion | Chr3 | 7907665 | 7907666 | 2 | |
| Insertion | Chr4 | 31644812 | 31644812 | 1 | |
| Insertion | Chr6 | 18639219 | 18639219 | 1 | |
| Insertion | Chr7 | 24850981 | 24851043 | 63 | |
| Insertion | Chr8 | 22256365 | 22256365 | 1 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 4603632 | 4632230 | |
| Inversion | Chr3 | 4603635 | 4632232 | |
| Inversion | Chr1 | 16207692 | 16640442 | LOC_Os01g28930 |
| Inversion | Chr1 | 16207704 | 16640448 | LOC_Os01g28930 |
| Inversion | Chr5 | 25571137 | 25571486 | |
| Inversion | Chr1 | 28128354 | 28266668 | |
| Inversion | Chr1 | 28128357 | 28266679 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 10912548 | Chr5 | 5898812 | |
| Translocation | Chr9 | 10912878 | Chr5 | 15553704 | LOC_Os05g26810 |
| Translocation | Chr9 | 10912891 | Chr5 | 15213734 | |
| Translocation | Chr3 | 15238103 | Chr2 | 3401798 | |
| Translocation | Chr7 | 19913844 | Chr4 | 22060737 | LOC_Os04g36570 |
| Translocation | Chr7 | 19914517 | Chr4 | 22060746 | 2 |
