Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3222-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3222-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 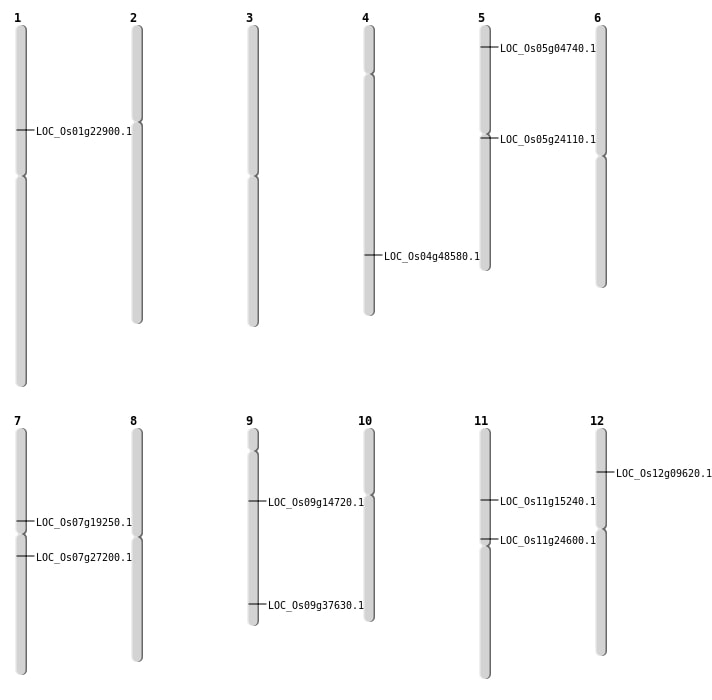
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19718518 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 42461969 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 15468601 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 19621380 | G-A | HET | INTRON | |
| SBS | Chr10 | 7192246 | G-C | HET | INTRON | |
| SBS | Chr11 | 13255409 | G-A | HET | INTRON | |
| SBS | Chr11 | 14038567 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g24600 |
| SBS | Chr11 | 25098877 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 7129008 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 8602958 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15240 |
| SBS | Chr11 | 9650770 | C-A | HET | INTRON | |
| SBS | Chr12 | 12994990 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 456549 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 9015428 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 13413931 | A-G | HET | INTRON | |
| SBS | Chr2 | 1897949 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 1897950 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 22322572 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 7323268 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 8340865 | G-T | HET | INTRON | |
| SBS | Chr2 | 8352695 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 32306243 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 35561766 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 5732480 | C-T | HET | INTRON | |
| SBS | Chr4 | 12911663 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 13753597 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 20918793 | T-G | HET | INTRON | |
| SBS | Chr4 | 28953747 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g48580 |
| SBS | Chr4 | 32139295 | T-G | HET | UTR_5_PRIME | |
| SBS | Chr4 | 34572440 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 34722443 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 8234687 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 8664270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 14675875 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 1595897 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 20092731 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 23772408 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 24200660 | C-T | HET | INTRON | |
| SBS | Chr6 | 26262835 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 4544642 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 7518997 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 15810734 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27200 |
| SBS | Chr7 | 27301553 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 8226557 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 8404042 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13155388 | C-G | HET | INTRON | |
| SBS | Chr8 | 19928606 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 8193004 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 21692505 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g37630 |
| SBS | Chr9 | 8101543 | G-A | HET | INTERGENIC |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1349112 | 1349114 | 2 | |
| Deletion | Chr5 | 4011866 | 4011867 | 1 | |
| Deletion | Chr3 | 6277322 | 6277323 | 1 | |
| Deletion | Chr9 | 8742002 | 8742003 | 1 | LOC_Os09g14720 |
| Deletion | Chr3 | 9418924 | 9418925 | 1 | |
| Deletion | Chr4 | 11285643 | 11285648 | 5 | |
| Deletion | Chr7 | 11399654 | 11399830 | 176 | LOC_Os07g19250 |
| Deletion | Chr8 | 11497741 | 11497795 | 54 | |
| Deletion | Chr3 | 14776801 | 14776802 | 1 | |
| Deletion | Chr9 | 18623957 | 18623980 | 23 | |
| Deletion | Chr10 | 19243724 | 19243725 | 1 | |
| Deletion | Chr5 | 22687618 | 22687624 | 6 | |
| Deletion | Chr5 | 23842832 | 23842838 | 6 |
Insertions: 10
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1965034 | 2115441 | |
| Inversion | Chr7 | 1965036 | 2115441 | |
| Inversion | Chr11 | 7778548 | 8668795 | |
| Inversion | Chr11 | 8653901 | 8668692 | |
| Inversion | Chr1 | 12871837 | 12871949 | LOC_Os01g22900 |
| Inversion | Chr8 | 16542493 | 16551207 | |
| Inversion | Chr8 | 16542494 | 16551208 | |
| Inversion | Chr6 | 30228545 | 30228796 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1294169 | Chr5 | 13907391 | LOC_Os05g24110 |
| Translocation | Chr10 | 1294176 | Chr5 | 13907553 | |
| Translocation | Chr2 | 4042426 | Chr1 | 7147934 | |
| Translocation | Chr2 | 4042890 | Chr1 | 5951819 | |
| Translocation | Chr12 | 4925533 | Chr6 | 1603851 | |
| Translocation | Chr12 | 5071425 | Chr6 | 1439554 | LOC_Os12g09620 |
| Translocation | Chr11 | 9031433 | Chr7 | 23098250 | |
| Translocation | Chr6 | 19273782 | Chr5 | 2247282 | LOC_Os05g04740 |
