Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3223-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3223-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 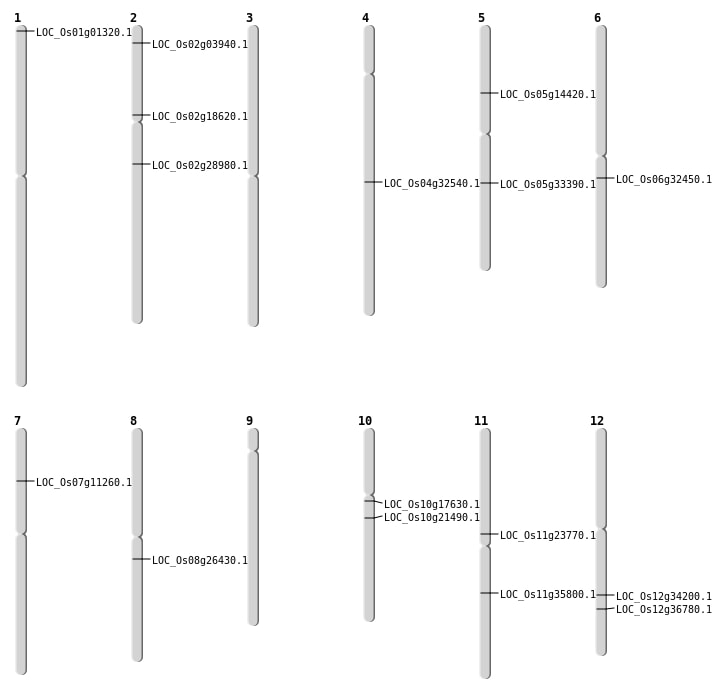
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14371578 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 21881050 | C-T | HET | INTRON | |
| SBS | Chr1 | 32568177 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 11007445 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g21490 |
| SBS | Chr11 | 13667642 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 14491803 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15827296 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14974726 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 15790017 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 20732103 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g34200 |
| SBS | Chr12 | 22550206 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g36780 |
| SBS | Chr12 | 23130077 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 26666147 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 9402093 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 15245889 | G-A | HET | INTRON | |
| SBS | Chr2 | 16294121 | C-T | HET | INTRON | |
| SBS | Chr2 | 1673299 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g03940 |
| SBS | Chr2 | 7073843 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 34499168 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 19612423 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g33390 |
| SBS | Chr6 | 10955644 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 22635786 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 13248267 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 14221291 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 5755682 | C-T | HET | INTRON | |
| SBS | Chr7 | 6210212 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11260 |
| SBS | Chr8 | 16104393 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g26430 |
| SBS | Chr8 | 19475034 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 366206 | A-T | HOMO | INTRON | |
| SBS | Chr8 | 4755255 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 17894609 | C-T | HET | INTRON | |
| SBS | Chr9 | 18554450 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 19012395 | G-A | HET | INTRON |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 157779 | 157809 | 30 | LOC_Os01g01320 |
| Deletion | Chr7 | 2154992 | 2155009 | 17 | |
| Deletion | Chr2 | 2604297 | 2604298 | 1 | |
| Deletion | Chr11 | 4527909 | 4527910 | 1 | |
| Deletion | Chr3 | 5197126 | 5197127 | 1 | |
| Deletion | Chr7 | 6283230 | 6283232 | 2 | |
| Deletion | Chr5 | 6371577 | 6371579 | 2 | |
| Deletion | Chr8 | 6973838 | 6973839 | 1 | |
| Deletion | Chr5 | 8134150 | 8134161 | 11 | LOC_Os05g14420 |
| Deletion | Chr7 | 8953033 | 8953035 | 2 | |
| Deletion | Chr6 | 10755062 | 10755063 | 1 | |
| Deletion | Chr2 | 13432504 | 13432507 | 3 | |
| Deletion | Chr3 | 15426830 | 15426831 | 1 | |
| Deletion | Chr1 | 15553571 | 15553594 | 23 | |
| Deletion | Chr3 | 15793848 | 15793849 | 1 | |
| Deletion | Chr9 | 16799034 | 16799046 | 12 | |
| Deletion | Chr11 | 18081494 | 18081495 | 1 | |
| Deletion | Chr6 | 18637639 | 18637640 | 1 | |
| Deletion | Chr6 | 18870148 | 18870149 | 1 | LOC_Os06g32450 |
| Deletion | Chr4 | 19593521 | 19593527 | 6 | LOC_Os04g32540 |
| Deletion | Chr11 | 21013962 | 21013963 | 1 | LOC_Os11g35800 |
| Deletion | Chr3 | 22820351 | 22820357 | 6 | |
| Deletion | Chr5 | 28886672 | 28886673 | 1 | |
| Deletion | Chr6 | 30632753 | 30632754 | 1 | |
| Deletion | Chr1 | 38427405 | 38427406 | 1 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 158016 | 158016 | 1 | LOC_Os01g01320 |
| Insertion | Chr10 | 15203369 | 15203370 | 2 | |
| Insertion | Chr12 | 13318573 | 13318573 | 1 | |
| Insertion | Chr12 | 16949077 | 16949077 | 1 | |
| Insertion | Chr2 | 13029347 | 13029347 | 1 | |
| Insertion | Chr4 | 30507134 | 30507137 | 4 | |
| Insertion | Chr5 | 5890607 | 5890607 | 1 | |
| Insertion | Chr6 | 25066084 | 25066084 | 1 | |
| Insertion | Chr8 | 2994270 | 2994271 | 2 | |
| Insertion | Chr8 | 7844749 | 7844749 | 1 | |
| Insertion | Chr9 | 14182830 | 14182831 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 8908912 | 8909125 | LOC_Os10g17630 |
| Inversion | Chr11 | 13247278 | 13445197 | LOC_Os11g23770 |
| Inversion | Chr11 | 13247288 | 13445271 | LOC_Os11g23770 |
| Inversion | Chr2 | 17156429 | 17156627 | LOC_Os02g28980 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2005847 | Chr2 | 19680995 | |
| Translocation | Chr5 | 3174844 | Chr2 | 13584625 | |
| Translocation | Chr11 | 4845284 | Chr4 | 30958425 | |
| Translocation | Chr5 | 5212055 | Chr1 | 26217213 | |
| Translocation | Chr11 | 5267295 | Chr5 | 13676750 | |
| Translocation | Chr12 | 10883302 | Chr2 | 25322026 | |
| Translocation | Chr12 | 12183087 | Chr2 | 9649794 | |
| Translocation | Chr12 | 19211759 | Chr2 | 10855496 | LOC_Os02g18620 |
| Translocation | Chr11 | 19842605 | Chr1 | 19348949 | |
| Translocation | Chr8 | 26299548 | Chr2 | 25044491 |
