Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3227-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3227-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 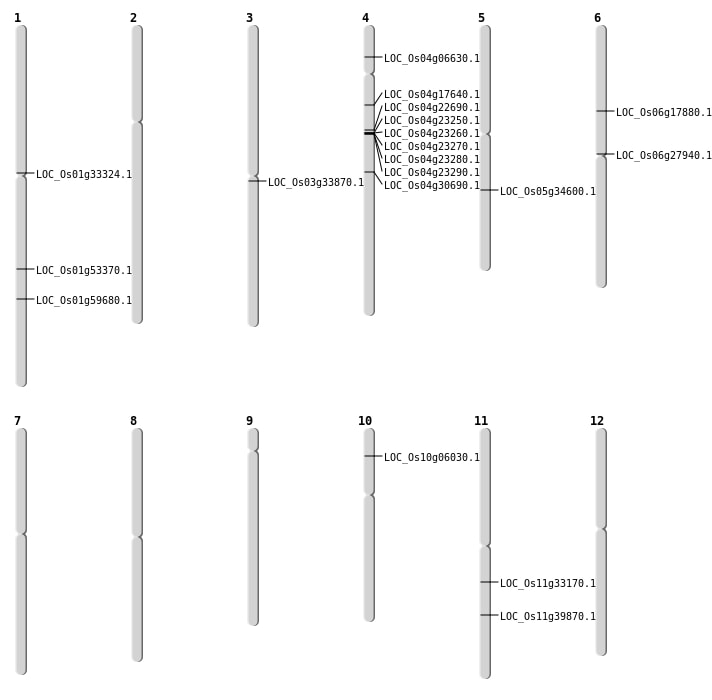
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1906402 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 30666817 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g53370 |
| SBS | Chr10 | 19564731 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 3064940 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g06030 |
| SBS | Chr11 | 15028448 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15882963 | G-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 21132182 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 5387063 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 11875546 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 26428094 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 3140797 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 12647073 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 35278849 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 3560791 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 1018875 | G-C | HET | INTRON | |
| SBS | Chr3 | 7251319 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 12862478 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22690 |
| SBS | Chr4 | 15212919 | C-G | HET | INTRON | |
| SBS | Chr4 | 9657809 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g17640 |
| SBS | Chr5 | 10776503 | T-G | HET | INTRON | |
| SBS | Chr5 | 16564419 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 20516812 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g34600 |
| SBS | Chr5 | 28155595 | T-C | HET | INTRON | |
| SBS | Chr5 | 28594647 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 8289515 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 9221214 | G-A | HET | INTRON | |
| SBS | Chr6 | 10378122 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g17880 |
| SBS | Chr6 | 12973705 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 16904080 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 6784935 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 12102181 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 15010427 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 16166952 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 16166957 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 19530881 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 28508474 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 28508475 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 9371450 | G-A | HET | INTRON | |
| SBS | Chr8 | 20902442 | A-G | HOMO | INTRON | |
| SBS | Chr9 | 3501268 | T-C | HOMO | INTRON | |
| SBS | Chr9 | 836282 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 194579 | 194582 | 3 | |
| Deletion | Chr10 | 2646609 | 2646610 | 1 | |
| Deletion | Chr9 | 2835769 | 2835770 | 1 | |
| Deletion | Chr12 | 4407609 | 4407610 | 1 | |
| Deletion | Chr11 | 7920716 | 7920717 | 1 | |
| Deletion | Chr8 | 9528777 | 9528778 | 1 | |
| Deletion | Chr10 | 10128690 | 10128691 | 1 | |
| Deletion | Chr1 | 11884277 | 11884279 | 2 | |
| Deletion | Chr4 | 13252001 | 13296000 | 43999 | 6 |
| Deletion | Chr9 | 13431388 | 13431396 | 8 | |
| Deletion | Chr5 | 15826728 | 15826729 | 1 | |
| Deletion | Chr6 | 15853422 | 15853434 | 12 | LOC_Os06g27940 |
| Deletion | Chr1 | 18359635 | 18359638 | 3 | LOC_Os01g33324 |
| Deletion | Chr11 | 18786678 | 18786685 | 7 | |
| Deletion | Chr11 | 19629376 | 19629385 | 9 | LOC_Os11g33170 |
| Deletion | Chr11 | 20200604 | 20200605 | 1 | |
| Deletion | Chr1 | 20352373 | 20352375 | 2 | |
| Deletion | Chr11 | 23768115 | 23768116 | 1 | LOC_Os11g39870 |
| Deletion | Chr6 | 26764325 | 26764326 | 1 | |
| Deletion | Chr11 | 27810238 | 27810239 | 1 | |
| Deletion | Chr1 | 28613305 | 28613307 | 2 | |
| Deletion | Chr2 | 31266368 | 31266370 | 2 | |
| Deletion | Chr3 | 34595815 | 34595822 | 7 | |
| Deletion | Chr1 | 39690288 | 39690289 | 1 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16269319 | 16269319 | 1 | |
| Insertion | Chr1 | 27819413 | 27819438 | 26 | |
| Insertion | Chr1 | 34932501 | 34932501 | 1 | |
| Insertion | Chr1 | 871582 | 871639 | 58 | |
| Insertion | Chr11 | 22688678 | 22688710 | 33 | |
| Insertion | Chr11 | 8175197 | 8175202 | 6 | |
| Insertion | Chr2 | 26793248 | 26793262 | 15 | |
| Insertion | Chr2 | 6757799 | 6757800 | 2 | |
| Insertion | Chr4 | 3491684 | 3491688 | 5 | LOC_Os04g06630 |
| Insertion | Chr5 | 24227394 | 24227413 | 20 | |
| Insertion | Chr7 | 8585877 | 8585877 | 1 | |
| Insertion | Chr8 | 7447113 | 7447113 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6291659 | 6747853 | |
| Inversion | Chr4 | 6291663 | 6747855 | |
| Inversion | Chr3 | 19055489 | 19348430 | LOC_Os03g33870 |
| Inversion | Chr3 | 19055500 | 19348440 | LOC_Os03g33870 |
| Inversion | Chr8 | 25364205 | 25365161 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 8402035 | Chr6 | 13377163 | |
| Translocation | Chr6 | 10457321 | Chr1 | 34461752 | |
| Translocation | Chr6 | 10457347 | Chr1 | 34526468 | LOC_Os01g59680 |
| Translocation | Chr12 | 18527485 | Chr11 | 14827073 | |
| Translocation | Chr8 | 24077241 | Chr4 | 20494294 |
