Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3228-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3228-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 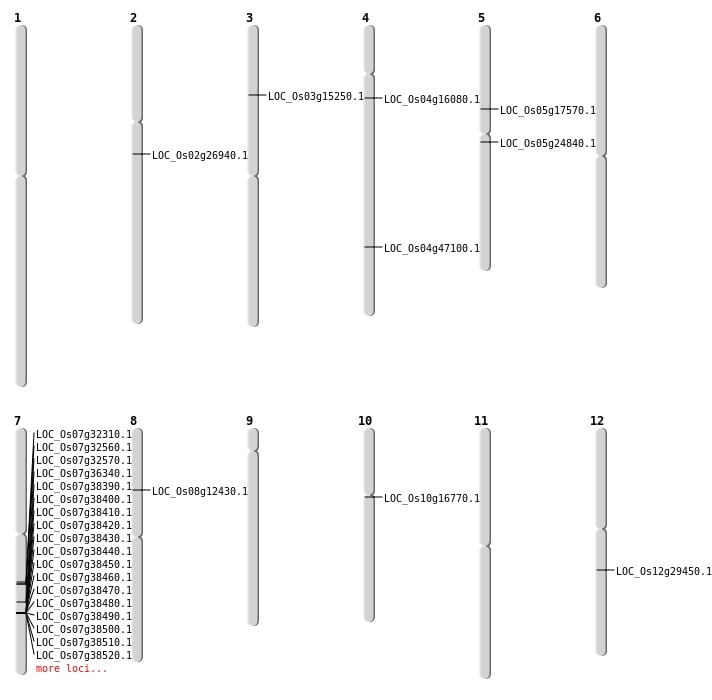
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10016629 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2999270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 35459776 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40679374 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 10946337 | G-T | HET | INTRON | |
| SBS | Chr10 | 9231320 | C-A | HET | INTRON | |
| SBS | Chr11 | 1205572 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 16020044 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 16205927 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr11 | 20955601 | C-T | HET | INTRON | |
| SBS | Chr11 | 21372520 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 21372521 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 21372533 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 8265186 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9903142 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 9903143 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 10034894 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 13705813 | G-A | HET | INTRON | |
| SBS | Chr12 | 17497073 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29450 |
| SBS | Chr12 | 19477213 | C-T | HET | INTRON | |
| SBS | Chr12 | 2137239 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 16698134 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 26725779 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 4811690 | G-A | HET | INTRON | |
| SBS | Chr2 | 8075244 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 3337615 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 33879838 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 7782174 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 12767969 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 24572535 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 27967253 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g47100 |
| SBS | Chr4 | 6012452 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 8716368 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16080 |
| SBS | Chr5 | 10094283 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17570 |
| SBS | Chr5 | 11964346 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 18876491 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 26461044 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 6684760 | A-T | HET | INTRON | |
| SBS | Chr6 | 7668628 | T-A | HET | INTRON | |
| SBS | Chr7 | 15136860 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 19213121 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32310 |
| SBS | Chr7 | 19345384 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 12347053 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 1805736 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 22283208 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 4781242 | C-T | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1430159 | 1430163 | 4 | |
| Deletion | Chr2 | 2359559 | 2359560 | 1 | |
| Deletion | Chr12 | 4830377 | 4830379 | 2 | |
| Deletion | Chr8 | 6378823 | 6378824 | 1 | |
| Deletion | Chr7 | 7013246 | 7013256 | 10 | |
| Deletion | Chr3 | 7667855 | 7667856 | 1 | |
| Deletion | Chr3 | 8340303 | 8340379 | 76 | LOC_Os03g15250 |
| Deletion | Chr10 | 8355110 | 8355113 | 3 | LOC_Os10g16770 |
| Deletion | Chr1 | 8469463 | 8469483 | 20 | |
| Deletion | Chr6 | 8536355 | 8536356 | 1 | |
| Deletion | Chr8 | 10085791 | 10085799 | 8 | |
| Deletion | Chr1 | 10810839 | 10810840 | 1 | |
| Deletion | Chr3 | 13184999 | 13185011 | 12 | |
| Deletion | Chr5 | 14397987 | 14397995 | 8 | LOC_Os05g24840 |
| Deletion | Chr7 | 19395001 | 19415000 | 19999 | 3 |
| Deletion | Chr6 | 19480050 | 19480051 | 1 | |
| Deletion | Chr11 | 20063236 | 20063238 | 2 | |
| Deletion | Chr1 | 20424456 | 20424462 | 6 | |
| Deletion | Chr6 | 22253964 | 22253965 | 1 | |
| Deletion | Chr7 | 23075001 | 23191000 | 115999 | 23 |
| Deletion | Chr11 | 27072233 | 27072234 | 1 | |
| Deletion | Chr6 | 30381774 | 30381777 | 3 |
Insertions: 6
No Inversion
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 4603985 | Chr1 | 12053283 | |
| Translocation | Chr8 | 4604015 | Chr2 | 15824153 | 2 |
| Translocation | Chr8 | 7339095 | Chr7 | 23192091 | LOC_Os08g12430 |
| Translocation | Chr8 | 7339097 | Chr1 | 12054429 | LOC_Os08g12430 |
| Translocation | Chr12 | 10448829 | Chr7 | 25514893 | |
| Translocation | Chr2 | 13261358 | Chr1 | 42763711 | |
| Translocation | Chr8 | 14590229 | Chr3 | 9391830 | |
| Translocation | Chr12 | 15119303 | Chr8 | 12191012 | |
| Translocation | Chr8 | 24322448 | Chr4 | 7832127 | |
| Translocation | Chr8 | 24322462 | Chr4 | 7831862 |
