Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3248-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3248-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 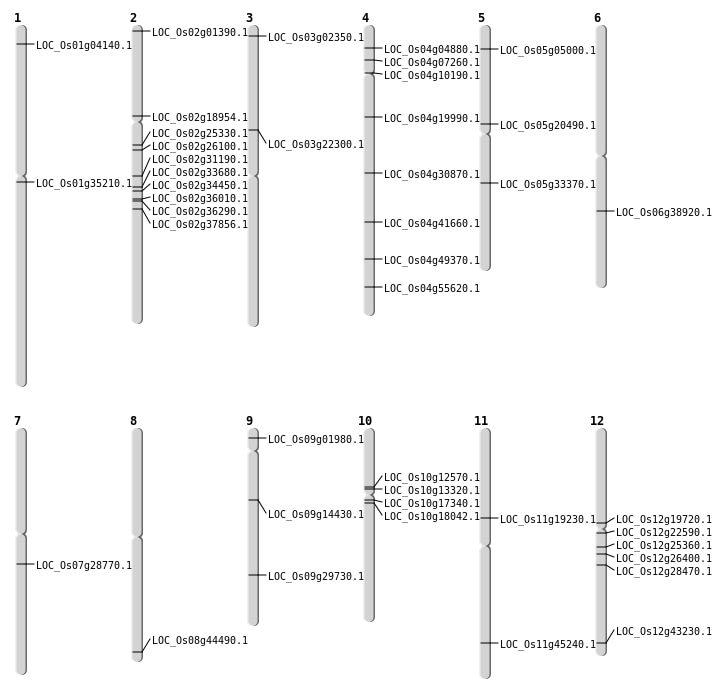
|
| Variant Information |
|---|
Single base substitutions: 18
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25418134 | C-A | HOMO | INTRON | |
| SBS | Chr10 | 12361220 | C-T | HET | INTRON | |
| SBS | Chr11 | 518555 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 8239495 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 12068757 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 16828399 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28470 |
| SBS | Chr2 | 15336760 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g26100 |
| SBS | Chr2 | 22872844 | T-A | HET | STOP_GAINED | LOC_Os02g37856 |
| SBS | Chr2 | 31095157 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 34533100 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 9810243 | A-G | HOMO | INTRON | |
| SBS | Chr3 | 29321548 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 6882163 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 5188344 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 120252 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 16850956 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 27982629 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g44490 |
| SBS | Chr9 | 8530445 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g14430 |
Deletions: 125
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 30197 | 30549 | 352 | |
| Deletion | Chr9 | 106441 | 106442 | 1 | |
| Deletion | Chr2 | 212723 | 212725 | 2 | |
| Deletion | Chr2 | 233169 | 233170 | 1 | LOC_Os02g01390 |
| Deletion | Chr5 | 682769 | 682770 | 1 | |
| Deletion | Chr3 | 1392150 | 1392152 | 2 | |
| Deletion | Chr8 | 1444754 | 1444755 | 1 | |
| Deletion | Chr1 | 1820204 | 1820207 | 3 | LOC_Os01g04140 |
| Deletion | Chr9 | 1946593 | 1946594 | 1 | |
| Deletion | Chr1 | 2008374 | 2008375 | 1 | |
| Deletion | Chr2 | 2049139 | 2049142 | 3 | |
| Deletion | Chr8 | 2049740 | 2049741 | 1 | |
| Deletion | Chr4 | 2633535 | 2633536 | 1 | |
| Deletion | Chr2 | 2707908 | 2707979 | 71 | |
| Deletion | Chr4 | 3362036 | 3362038 | 2 | |
| Deletion | Chr10 | 3386842 | 3386908 | 66 | |
| Deletion | Chr3 | 3707272 | 3707273 | 1 | |
| Deletion | Chr5 | 3997276 | 3997277 | 1 | |
| Deletion | Chr8 | 4370741 | 4370742 | 1 | |
| Deletion | Chr9 | 4517139 | 4517140 | 1 | |
| Deletion | Chr5 | 4521173 | 4521174 | 1 | |
| Deletion | Chr1 | 4787551 | 4787552 | 1 | |
| Deletion | Chr10 | 5024451 | 5024480 | 29 | |
| Deletion | Chr8 | 5195722 | 5195723 | 1 | |
| Deletion | Chr4 | 5348461 | 5348465 | 4 | |
| Deletion | Chr5 | 5438040 | 5438042 | 2 | |
| Deletion | Chr4 | 5455958 | 5455959 | 1 | |
| Deletion | Chr8 | 5495747 | 5495748 | 1 | |
| Deletion | Chr4 | 5510393 | 5510411 | 18 | LOC_Os04g10190 |
| Deletion | Chr9 | 5754991 | 5754992 | 1 | |
| Deletion | Chr7 | 6159345 | 6159346 | 1 | |
| Deletion | Chr5 | 6620034 | 6620060 | 26 | |
| Deletion | Chr10 | 6821333 | 6821334 | 1 | |
| Deletion | Chr10 | 7004734 | 7004737 | 3 | LOC_Os10g12570 |
| Deletion | Chr10 | 7190781 | 7190784 | 3 | LOC_Os10g13320 |
| Deletion | Chr5 | 7540094 | 7540112 | 18 | |
| Deletion | Chr1 | 7898139 | 7898221 | 82 | |
| Deletion | Chr5 | 7898988 | 7898990 | 2 | |
| Deletion | Chr10 | 8096360 | 8096450 | 90 | |
| Deletion | Chr10 | 9142666 | 9142667 | 1 | |
| Deletion | Chr10 | 9143935 | 9143938 | 3 | LOC_Os10g18042 |
| Deletion | Chr8 | 9162053 | 9162066 | 13 | |
| Deletion | Chr11 | 10024566 | 10024568 | 2 | |
| Deletion | Chr7 | 10836465 | 10836466 | 1 | |
| Deletion | Chr10 | 10959300 | 10959301 | 1 | |
| Deletion | Chr11 | 10995148 | 10995150 | 2 | LOC_Os11g19230 |
| Deletion | Chr4 | 11145276 | 11145279 | 3 | LOC_Os04g19990 |
| Deletion | Chr4 | 11152005 | 11152060 | 55 | |
| Deletion | Chr9 | 11328800 | 11328801 | 1 | |
| Deletion | Chr1 | 11398181 | 11398183 | 2 | |
| Deletion | Chr6 | 11425551 | 11425552 | 1 | |
| Deletion | Chr12 | 11487340 | 11487341 | 1 | LOC_Os12g19720 |
| Deletion | Chr5 | 11604566 | 11604567 | 1 | |
| Deletion | Chr8 | 11637046 | 11637062 | 16 | |
| Deletion | Chr1 | 12180234 | 12180235 | 1 | |
| Deletion | Chr1 | 12473482 | 12473483 | 1 | |
| Deletion | Chr4 | 12527674 | 12527675 | 1 | |
| Deletion | Chr5 | 12606746 | 12606747 | 1 | |
| Deletion | Chr12 | 12748236 | 12748262 | 26 | LOC_Os12g22590 |
| Deletion | Chr1 | 12951506 | 12951507 | 1 | |
| Deletion | Chr8 | 13096380 | 13096381 | 1 | |
| Deletion | Chr8 | 13578865 | 13578917 | 52 | |
| Deletion | Chr4 | 13728895 | 13728896 | 1 | 2 |
| Deletion | Chr6 | 13770352 | 13770354 | 2 | |
| Deletion | Chr8 | 13799011 | 13799012 | 1 | |
| Deletion | Chr8 | 13807996 | 13807997 | 1 | |
| Deletion | Chr7 | 14012509 | 14012511 | 2 | |
| Deletion | Chr9 | 14121999 | 14122001 | 2 | |
| Deletion | Chr1 | 14254043 | 14254074 | 31 | |
| Deletion | Chr5 | 14559802 | 14559803 | 1 | |
| Deletion | Chr7 | 14974925 | 14974926 | 1 | |
| Deletion | Chr8 | 15298518 | 15298519 | 1 | |
| Deletion | Chr12 | 15425789 | 15425790 | 1 | LOC_Os12g26400 |
| Deletion | Chr4 | 15697413 | 15697424 | 11 | |
| Deletion | Chr11 | 16017827 | 16017828 | 1 | |
| Deletion | Chr1 | 16090567 | 16090570 | 3 | |
| Deletion | Chr12 | 16274661 | 16274662 | 1 | |
| Deletion | Chr9 | 16373604 | 16373605 | 1 | |
| Deletion | Chr11 | 16518111 | 16518112 | 1 | |
| Deletion | Chr11 | 16553034 | 16553037 | 3 | |
| Deletion | Chr1 | 16644005 | 16644008 | 3 | |
| Deletion | Chr11 | 16715081 | 16715085 | 4 | |
| Deletion | Chr6 | 16760304 | 16760307 | 3 | |
| Deletion | Chr1 | 16773079 | 16773080 | 1 | |
| Deletion | Chr10 | 16889108 | 16889109 | 1 | |
| Deletion | Chr8 | 17025543 | 17025544 | 1 | |
| Deletion | Chr3 | 17308988 | 17308989 | 1 | |
| Deletion | Chr8 | 17546388 | 17546389 | 1 | |
| Deletion | Chr3 | 17955681 | 17955747 | 66 | |
| Deletion | Chr9 | 18079311 | 18079334 | 23 | LOC_Os09g29730 |
| Deletion | Chr9 | 18081938 | 18081939 | 1 | |
| Deletion | Chr4 | 18433001 | 18450000 | 16999 | 2 |
| Deletion | Chr2 | 18944577 | 18944578 | 1 | |
| Deletion | Chr1 | 19487736 | 19487809 | 73 | |
| Deletion | Chr12 | 19501223 | 19501225 | 2 | |
| Deletion | Chr1 | 19586970 | 19586971 | 1 | |
| Deletion | Chr5 | 19604027 | 19604028 | 1 | LOC_Os05g33370 |
| Deletion | Chr12 | 19773460 | 19773462 | 2 | |
| Deletion | Chr5 | 19861395 | 19861396 | 1 | |
| Deletion | Chr2 | 20642518 | 20642519 | 1 | LOC_Os02g34450 |
| Deletion | Chr11 | 20914642 | 20914643 | 1 | |
| Deletion | Chr5 | 21031185 | 21031256 | 71 | |
| Deletion | Chr12 | 21069314 | 21069315 | 1 | |
| Deletion | Chr11 | 21225747 | 21225749 | 2 | |
| Deletion | Chr7 | 21452372 | 21452373 | 1 | |
| Deletion | Chr2 | 21628696 | 21628698 | 2 | LOC_Os02g36010 |
| Deletion | Chr8 | 22073984 | 22073985 | 1 | |
| Deletion | Chr11 | 22392417 | 22392418 | 1 | |
| Deletion | Chr6 | 22790212 | 22790214 | 2 | |
| Deletion | Chr4 | 24701280 | 24701284 | 4 | LOC_Os04g41660 |
| Deletion | Chr6 | 25377805 | 25377810 | 5 | |
| Deletion | Chr1 | 26382072 | 26382073 | 1 | |
| Deletion | Chr12 | 26403195 | 26403197 | 2 | |
| Deletion | Chr5 | 26433924 | 26433927 | 3 | |
| Deletion | Chr12 | 26542753 | 26542754 | 1 | |
| Deletion | Chr12 | 26832904 | 26832905 | 1 | LOC_Os12g43230 |
| Deletion | Chr11 | 27379330 | 27379339 | 9 | LOC_Os11g45240 |
| Deletion | Chr3 | 27528813 | 27528829 | 16 | |
| Deletion | Chr3 | 28218060 | 28218061 | 1 | |
| Deletion | Chr4 | 28946449 | 28946450 | 1 | |
| Deletion | Chr5 | 29180296 | 29180302 | 6 | |
| Deletion | Chr6 | 30156123 | 30156125 | 2 | |
| Deletion | Chr4 | 31709157 | 31709158 | 1 | |
| Deletion | Chr4 | 32262548 | 32262581 | 33 | |
| Deletion | Chr4 | 35228248 | 35228249 | 1 |
Insertions: 107
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 13741222 | 13741222 | 1 | |
| Insertion | Chr1 | 13765384 | 13765385 | 2 | |
| Insertion | Chr1 | 15748933 | 15748933 | 1 | |
| Insertion | Chr1 | 16852109 | 16852116 | 8 | |
| Insertion | Chr1 | 19490438 | 19490440 | 3 | LOC_Os01g35210 |
| Insertion | Chr1 | 19523854 | 19523857 | 4 | |
| Insertion | Chr1 | 20487038 | 20487038 | 1 | |
| Insertion | Chr1 | 21327368 | 21327368 | 1 | |
| Insertion | Chr1 | 23420341 | 23420341 | 1 | |
| Insertion | Chr1 | 27444412 | 27444412 | 1 | |
| Insertion | Chr1 | 31465503 | 31465503 | 1 | |
| Insertion | Chr1 | 36853199 | 36853200 | 2 | |
| Insertion | Chr1 | 36889276 | 36889277 | 2 | |
| Insertion | Chr1 | 3794628 | 3794629 | 2 | |
| Insertion | Chr10 | 1254945 | 1254945 | 1 | |
| Insertion | Chr10 | 12849436 | 12849437 | 2 | |
| Insertion | Chr10 | 15179776 | 15179776 | 1 | |
| Insertion | Chr10 | 5104974 | 5104974 | 1 | |
| Insertion | Chr10 | 7181276 | 7181277 | 2 | |
| Insertion | Chr10 | 8733041 | 8733041 | 1 | LOC_Os10g17340 |
| Insertion | Chr11 | 20201694 | 20201698 | 5 | |
| Insertion | Chr11 | 27830603 | 27830606 | 4 | |
| Insertion | Chr11 | 28585656 | 28585689 | 34 | |
| Insertion | Chr11 | 9979902 | 9979902 | 1 | |
| Insertion | Chr12 | 12111714 | 12111714 | 1 | |
| Insertion | Chr12 | 12616516 | 12616516 | 1 | |
| Insertion | Chr12 | 14632611 | 14632637 | 27 | LOC_Os12g25360 |
| Insertion | Chr12 | 18144669 | 18144669 | 1 | |
| Insertion | Chr12 | 20407378 | 20407382 | 5 | |
| Insertion | Chr12 | 21603178 | 21603210 | 33 | |
| Insertion | Chr12 | 23365140 | 23365143 | 4 | |
| Insertion | Chr12 | 25786377 | 25786377 | 1 | |
| Insertion | Chr12 | 5322117 | 5322117 | 1 | |
| Insertion | Chr12 | 7861029 | 7861029 | 1 | |
| Insertion | Chr12 | 9721026 | 9721026 | 1 | |
| Insertion | Chr2 | 12254823 | 12254824 | 2 | |
| Insertion | Chr2 | 13479110 | 13479128 | 19 | |
| Insertion | Chr2 | 14767637 | 14767637 | 1 | LOC_Os02g25330 |
| Insertion | Chr2 | 18680854 | 18680854 | 1 | LOC_Os02g31190 |
| Insertion | Chr2 | 19722828 | 19722834 | 7 | |
| Insertion | Chr2 | 21712057 | 21712058 | 2 | |
| Insertion | Chr2 | 21913119 | 21913119 | 1 | LOC_Os02g36290 |
| Insertion | Chr2 | 7458562 | 7458566 | 5 | |
| Insertion | Chr2 | 9354444 | 9354444 | 1 | |
| Insertion | Chr3 | 13209063 | 13209063 | 1 | |
| Insertion | Chr3 | 18907122 | 18907123 | 2 | |
| Insertion | Chr3 | 19102222 | 19102225 | 4 | |
| Insertion | Chr3 | 2456232 | 2456237 | 6 | |
| Insertion | Chr3 | 28500708 | 28500718 | 11 | |
| Insertion | Chr3 | 35505770 | 35505770 | 1 | |
| Insertion | Chr3 | 36408469 | 36408470 | 2 | |
| Insertion | Chr3 | 823631 | 823634 | 4 | LOC_Os03g02350 |
| Insertion | Chr4 | 10491365 | 10491366 | 2 | |
| Insertion | Chr4 | 13728895 | 13728895 | 1 | 2 |
| Insertion | Chr4 | 14413256 | 14413256 | 1 | |
| Insertion | Chr4 | 1729130 | 1729131 | 2 | |
| Insertion | Chr4 | 20755843 | 20755843 | 1 | |
| Insertion | Chr4 | 2349290 | 2349290 | 1 | LOC_Os04g04880 |
| Insertion | Chr4 | 2724297 | 2724297 | 1 | |
| Insertion | Chr4 | 3005728 | 3005728 | 1 | |
| Insertion | Chr4 | 3272698 | 3272703 | 6 | |
| Insertion | Chr4 | 3853765 | 3853767 | 3 | LOC_Os04g07260 |
| Insertion | Chr5 | 10106087 | 10106087 | 1 | |
| Insertion | Chr5 | 11217752 | 11217752 | 1 | |
| Insertion | Chr5 | 12568685 | 12568753 | 69 | |
| Insertion | Chr5 | 15435989 | 15435992 | 4 | |
| Insertion | Chr5 | 19007447 | 19007513 | 67 | |
| Insertion | Chr5 | 2040198 | 2040198 | 1 | |
| Insertion | Chr5 | 2429594 | 2429594 | 1 | LOC_Os05g05000 |
| Insertion | Chr5 | 25602269 | 25602269 | 1 | |
| Insertion | Chr5 | 2808568 | 2808571 | 4 | |
| Insertion | Chr5 | 4954541 | 4954546 | 6 | |
| Insertion | Chr5 | 5593266 | 5593318 | 53 | |
| Insertion | Chr5 | 8194568 | 8194568 | 1 | |
| Insertion | Chr5 | 8774912 | 8774912 | 1 | |
| Insertion | Chr6 | 10294224 | 10294224 | 1 | |
| Insertion | Chr6 | 12518358 | 12518358 | 1 | |
| Insertion | Chr6 | 13904097 | 13904098 | 2 | |
| Insertion | Chr6 | 14042087 | 14042091 | 5 | |
| Insertion | Chr6 | 15083983 | 15083986 | 4 | |
| Insertion | Chr6 | 23090979 | 23090979 | 1 | LOC_Os06g38920 |
| Insertion | Chr6 | 29300028 | 29300058 | 31 | |
| Insertion | Chr7 | 16841968 | 16842003 | 36 | LOC_Os07g28770 |
| Insertion | Chr7 | 27597848 | 27597850 | 3 | |
| Insertion | Chr7 | 7461902 | 7461902 | 1 | |
| Insertion | Chr7 | 9802103 | 9802103 | 1 | |
| Insertion | Chr7 | 9954189 | 9954189 | 1 | |
| Insertion | Chr8 | 13595702 | 13595702 | 1 | |
| Insertion | Chr8 | 15818195 | 15818195 | 1 | |
| Insertion | Chr8 | 17507637 | 17507647 | 11 | |
| Insertion | Chr8 | 17582030 | 17582030 | 1 | |
| Insertion | Chr8 | 20957028 | 20957028 | 1 | |
| Insertion | Chr8 | 24362510 | 24362510 | 1 | |
| Insertion | Chr8 | 26662495 | 26662495 | 1 | |
| Insertion | Chr8 | 26718688 | 26718689 | 2 | |
| Insertion | Chr8 | 6938082 | 6938083 | 2 | |
| Insertion | Chr8 | 7266096 | 7266097 | 2 | |
| Insertion | Chr8 | 7313746 | 7313746 | 1 | |
| Insertion | Chr9 | 16373313 | 16373315 | 3 | |
| Insertion | Chr9 | 17785615 | 17785615 | 1 | |
| Insertion | Chr9 | 18633431 | 18633432 | 2 | |
| Insertion | Chr9 | 3286018 | 3286018 | 1 | |
| Insertion | Chr9 | 4855689 | 4855690 | 2 | |
| Insertion | Chr9 | 5354976 | 5354976 | 1 | |
| Insertion | Chr9 | 6695975 | 6695988 | 14 | |
| Insertion | Chr9 | 7426700 | 7426700 | 1 | |
| Insertion | Chr9 | 8474198 | 8474198 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 6641179 | 6655804 |
Translocations: 26
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 301058 | Chr3 | 12783338 | LOC_Os03g22300 |
| Translocation | Chr9 | 681291 | Chr3 | 24404315 | LOC_Os09g01980 |
| Translocation | Chr9 | 682399 | Chr2 | 11074026 | 2 |
| Translocation | Chr4 | 729097 | Chr3 | 31273134 | |
| Translocation | Chr4 | 2085445 | Chr1 | 27847846 | |
| Translocation | Chr9 | 6280172 | Chr6 | 20090155 | |
| Translocation | Chr7 | 8834473 | Chr5 | 10239419 | |
| Translocation | Chr12 | 8840829 | Chr9 | 20497358 | |
| Translocation | Chr3 | 9428792 | Chr1 | 16513483 | |
| Translocation | Chr7 | 9666167 | Chr4 | 14117299 | |
| Translocation | Chr12 | 9879033 | Chr10 | 327477 | |
| Translocation | Chr11 | 11559601 | Chr4 | 11340018 | |
| Translocation | Chr10 | 12685695 | Chr9 | 1170880 | |
| Translocation | Chr11 | 12810740 | Chr5 | 8349916 | |
| Translocation | Chr8 | 15300256 | Chr2 | 20643804 | LOC_Os02g34450 |
| Translocation | Chr11 | 15339318 | Chr3 | 15104040 | |
| Translocation | Chr10 | 15557603 | Chr4 | 15030348 | |
| Translocation | Chr11 | 18646856 | Chr2 | 19207247 | |
| Translocation | Chr10 | 19398743 | Chr8 | 19810927 | |
| Translocation | Chr11 | 20175274 | Chr2 | 20082593 | LOC_Os02g33680 |
| Translocation | Chr10 | 22644181 | Chr8 | 19749033 | |
| Translocation | Chr6 | 22703991 | Chr3 | 10562565 | |
| Translocation | Chr4 | 23032051 | Chr3 | 22887559 | |
| Translocation | Chr3 | 24403711 | Chr2 | 11073643 | LOC_Os02g18954 |
| Translocation | Chr7 | 27359823 | Chr5 | 12027449 | LOC_Os05g20490 |
| Translocation | Chr4 | 33111949 | Chr2 | 8784450 | LOC_Os04g55620 |
