Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3251-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3251-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 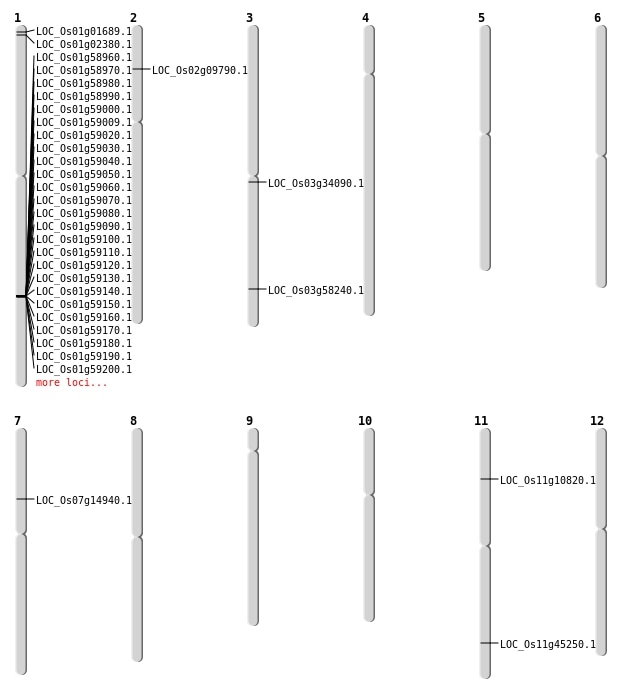
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29216559 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 31154623 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 31155066 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 37643000 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g64840 |
| SBS | Chr10 | 17931685 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 18652737 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 5972745 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 11917205 | C-T | HET | INTRON | |
| SBS | Chr11 | 11926971 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 14044299 | G-A | HET | INTRON | |
| SBS | Chr11 | 8533037 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 18234099 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 9721090 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 9721091 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 32005877 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 18789240 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19427196 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g34090 |
| SBS | Chr3 | 21020102 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr3 | 27766264 | G-A | HET | INTRON | |
| SBS | Chr3 | 31003895 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 34720459 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 11625701 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 19540367 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 2662877 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 2662878 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 27381212 | T-A | HET | INTRON | |
| SBS | Chr5 | 3210590 | A-C | HET | INTRON | |
| SBS | Chr6 | 13577251 | T-G | HET | INTRON | |
| SBS | Chr6 | 17693030 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 20803549 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 19809059 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8562236 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g14940 |
| SBS | Chr8 | 25303806 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 7293363 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 872750 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 1462236 | G-A | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 306066 | 306067 | 1 | |
| Deletion | Chr1 | 761447 | 761512 | 65 | LOC_Os01g02380 |
| Deletion | Chr2 | 1705357 | 1705368 | 11 | |
| Deletion | Chr11 | 2914001 | 2924000 | 9999 | LOC_Os11g10820 |
| Deletion | Chr10 | 3278568 | 3278569 | 1 | |
| Deletion | Chr8 | 5912934 | 5912942 | 8 | |
| Deletion | Chr9 | 7291693 | 7291790 | 97 | |
| Deletion | Chr4 | 7295544 | 7295550 | 6 | |
| Deletion | Chr4 | 8605174 | 8605218 | 44 | |
| Deletion | Chr9 | 9684023 | 9684024 | 1 | |
| Deletion | Chr6 | 9810369 | 9810376 | 7 | |
| Deletion | Chr9 | 11086218 | 11086265 | 47 | |
| Deletion | Chr2 | 11449152 | 11449153 | 1 | |
| Deletion | Chr8 | 12737280 | 12737283 | 3 | |
| Deletion | Chr9 | 15181692 | 15181720 | 28 | |
| Deletion | Chr3 | 16232252 | 16232316 | 64 | |
| Deletion | Chr9 | 16583962 | 16583971 | 9 | |
| Deletion | Chr12 | 17891911 | 17891917 | 6 | |
| Deletion | Chr12 | 18058519 | 18058520 | 1 | |
| Deletion | Chr8 | 20307392 | 20307438 | 46 | |
| Deletion | Chr12 | 21379901 | 21379902 | 1 | |
| Deletion | Chr4 | 23247001 | 23267000 | 19999 | LOC_Os01g01689 |
| Deletion | Chr1 | 25822942 | 25822990 | 48 | |
| Deletion | Chr12 | 26328755 | 26328756 | 1 | |
| Deletion | Chr11 | 27240768 | 27240771 | 3 | |
| Deletion | Chr1 | 34070001 | 34367000 | 296999 | 46 |
| Deletion | Chr1 | 41615885 | 41615886 | 1 |
Insertions: 14
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 4865403 | 5051634 | LOC_Os02g09790 |
| Inversion | Chr2 | 7702621 | 8081622 | |
| Inversion | Chr2 | 7702631 | 8081632 | |
| Inversion | Chr10 | 19162984 | 19308722 | |
| Inversion | Chr7 | 25245647 | 25245995 | |
| Inversion | Chr1 | 33863046 | 34070968 | |
| Inversion | Chr1 | 33866149 | 34382579 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 7057134 | Chr1 | 31953047 | |
| Translocation | Chr12 | 13966747 | Chr1 | 19217180 | |
| Translocation | Chr11 | 24413996 | Chr5 | 5542130 | |
| Translocation | Chr11 | 27384608 | Chr3 | 33184708 | LOC_Os11g45250 |
| Translocation | Chr11 | 27384802 | Chr3 | 33184707 | 2 |
