Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3253-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3253-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 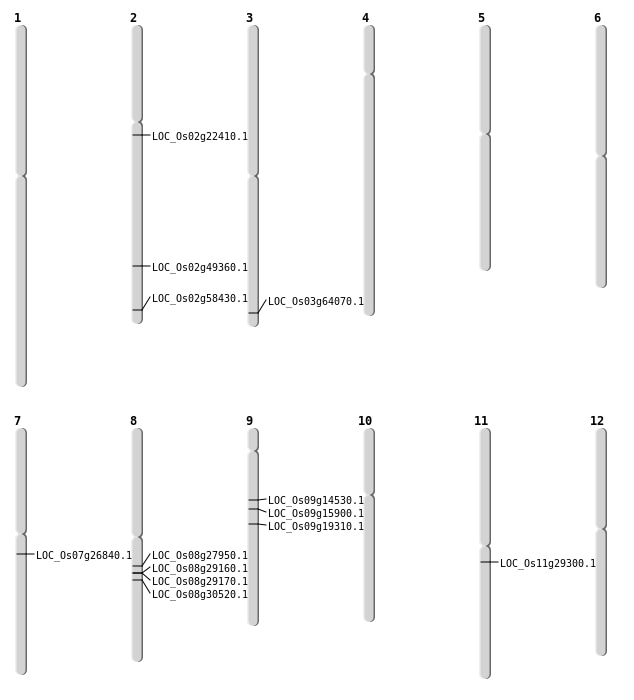
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16316277 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 26563573 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 41322729 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15545622 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 16992216 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29300 |
| SBS | Chr11 | 20284298 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 2097945 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 22363054 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 26817301 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 13848988 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 21241887 | T-C | HET | INTRON | |
| SBS | Chr12 | 24078426 | G-T | HET | INTRON | |
| SBS | Chr12 | 4711779 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 13386087 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g22410 |
| SBS | Chr2 | 30159574 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g49360 |
| SBS | Chr2 | 32398787 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35734030 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g58430 |
| SBS | Chr3 | 36204771 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g64070 |
| SBS | Chr4 | 10517118 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 13561004 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 12270422 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 12899884 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 15810741 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 28574769 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 2892028 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 15527973 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g26840 |
| SBS | Chr7 | 15527974 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g26840 |
| SBS | Chr7 | 23726717 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 24233367 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 26283700 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13085598 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 20638453 | G-A | HET | INTRON | |
| SBS | Chr9 | 3643453 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 8355839 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 8595496 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14530 |
| SBS | Chr9 | 9715269 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15900 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 194174 | 194186 | 12 | |
| Deletion | Chr8 | 467156 | 467186 | 30 | |
| Deletion | Chr6 | 1623801 | 1623811 | 10 | |
| Deletion | Chr10 | 2089659 | 2089662 | 3 | |
| Deletion | Chr12 | 4140902 | 4140903 | 1 | |
| Deletion | Chr1 | 6270151 | 6270170 | 19 | |
| Deletion | Chr7 | 7187008 | 7187021 | 13 | |
| Deletion | Chr3 | 10710998 | 10710999 | 1 | |
| Deletion | Chr3 | 10848548 | 10848549 | 1 | |
| Deletion | Chr10 | 13981781 | 13981801 | 20 | |
| Deletion | Chr6 | 14038562 | 14038563 | 1 | |
| Deletion | Chr1 | 14280361 | 14280371 | 10 | |
| Deletion | Chr6 | 15337485 | 15337486 | 1 | |
| Deletion | Chr8 | 17034909 | 17034917 | 8 | LOC_Os08g27950 |
| Deletion | Chr9 | 17554980 | 17554993 | 13 | |
| Deletion | Chr8 | 17856001 | 17866000 | 9999 | 3 |
| Deletion | Chr10 | 18716706 | 18716711 | 5 | |
| Deletion | Chr6 | 21466097 | 21466098 | 1 | |
| Deletion | Chr11 | 21760683 | 21760686 | 3 | |
| Deletion | Chr11 | 22237172 | 22237189 | 17 | |
| Deletion | Chr7 | 23156472 | 23156473 | 1 | |
| Deletion | Chr8 | 24762759 | 24762761 | 2 | |
| Deletion | Chr8 | 25006367 | 25006368 | 1 | |
| Deletion | Chr7 | 25355082 | 25355100 | 18 | |
| Deletion | Chr11 | 26698918 | 26698919 | 1 | |
| Deletion | Chr1 | 26819903 | 26819915 | 12 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 9987199 | 9987199 | 1 | |
| Insertion | Chr6 | 7423159 | 7423159 | 1 | |
| Insertion | Chr6 | 7457203 | 7457203 | 1 | |
| Insertion | Chr8 | 6385387 | 6385387 | 1 | |
| Insertion | Chr9 | 11555173 | 11555173 | 1 | LOC_Os09g19310 |
No Inversion
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 15461752 | Chr3 | 8299083 | |
| Translocation | Chr9 | 15462987 | Chr3 | 8299071 | |
| Translocation | Chr9 | 15536793 | Chr4 | 33688938 | |
| Translocation | Chr9 | 15537146 | Chr4 | 33688938 |
