Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3255-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3255-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 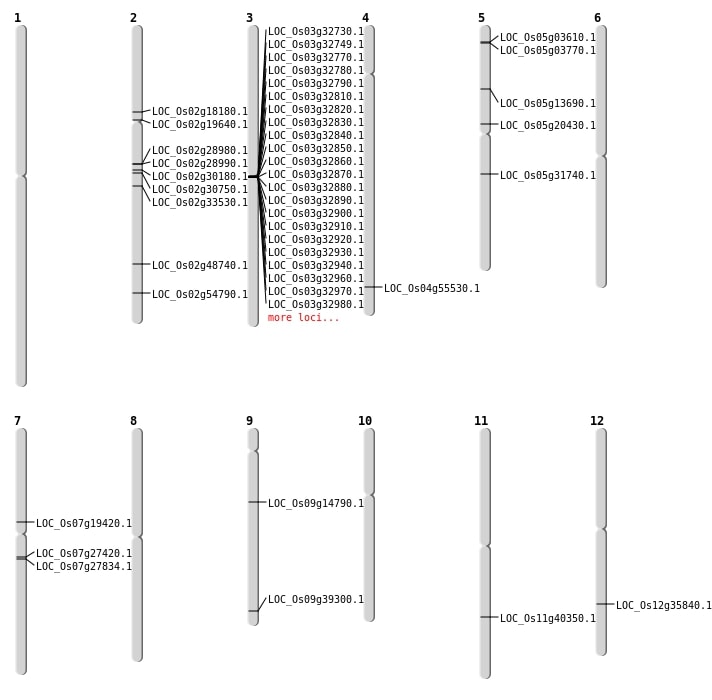
|
| Variant Information |
|---|
Single base substitutions: 72
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14258265 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 29554082 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 39328441 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7978743 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 1177952 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 12243258 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 12354746 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 14690929 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 5040174 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 581616 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 8076706 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 9122881 | A-C | HET | INTRON | |
| SBS | Chr11 | 10502003 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11140601 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11746296 | G-A | HET | INTRON | |
| SBS | Chr11 | 16015440 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21769306 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 23906724 | T-A | HET | INTRON | |
| SBS | Chr12 | 11096290 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 15598447 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 21841571 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g35840 |
| SBS | Chr2 | 10554914 | C-A | HET | STOP_GAINED | LOC_Os02g18180 |
| SBS | Chr2 | 10609064 | G-T | HET | INTRON | |
| SBS | Chr2 | 11478370 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g19640 |
| SBS | Chr2 | 14138355 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 18325912 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g30750 |
| SBS | Chr2 | 20212545 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 20696147 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 30040025 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 33552211 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g54790 |
| SBS | Chr2 | 33552212 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 5879219 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 8745235 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 12395140 | G-A | HET | INTRON | |
| SBS | Chr3 | 23108387 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31332151 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 34508821 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr3 | 35483191 | A-G | HET | INTRON | |
| SBS | Chr4 | 10267264 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 12788464 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 13138497 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 15257312 | C-T | HET | INTRON | |
| SBS | Chr4 | 25745769 | C-A | HET | INTRON | |
| SBS | Chr4 | 27676175 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 29964377 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 31894888 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 32468166 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 32943656 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 33037971 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g55530 |
| SBS | Chr5 | 11988951 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g20430 |
| SBS | Chr5 | 434157 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 7586624 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g13690 |
| SBS | Chr6 | 12762671 | A-G | HET | INTRON | |
| SBS | Chr6 | 14177545 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 14520551 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 22976226 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 25435764 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 2925663 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 640321 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr6 | 8746687 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 973127 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 11489776 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g19420 |
| SBS | Chr7 | 11874894 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 15977481 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27420 |
| SBS | Chr7 | 21128070 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 29019402 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 3320537 | A-T | HET | INTRON | |
| SBS | Chr7 | 9969733 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 3985460 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 4626694 | G-A | HET | INTRON | |
| SBS | Chr9 | 8801443 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14790 |
| SBS | Chr9 | 9676857 | G-T | HOMO | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 497425 | 497431 | 6 | |
| Deletion | Chr11 | 1204964 | 1204999 | 35 | |
| Deletion | Chr4 | 2185074 | 2185077 | 3 | |
| Deletion | Chr6 | 2260270 | 2260302 | 32 | |
| Deletion | Chr9 | 3332556 | 3333154 | 598 | |
| Deletion | Chr4 | 8947719 | 8947723 | 4 | |
| Deletion | Chr8 | 9574971 | 9574972 | 1 | |
| Deletion | Chr8 | 12006856 | 12006858 | 2 | |
| Deletion | Chr3 | 12313908 | 12313910 | 2 | |
| Deletion | Chr7 | 14544001 | 14565000 | 20999 | LOC_Os07g27834 |
| Deletion | Chr5 | 16460986 | 16460987 | 1 | |
| Deletion | Chr2 | 17148001 | 17165000 | 16999 | 3 |
| Deletion | Chr5 | 18488574 | 18488583 | 9 | LOC_Os05g31740 |
| Deletion | Chr3 | 18742001 | 18916000 | 173999 | 32 |
| Deletion | Chr2 | 19940927 | 19940928 | 1 | LOC_Os02g33530 |
| Deletion | Chr8 | 21324471 | 21324472 | 1 | |
| Deletion | Chr9 | 22587997 | 22588075 | 78 | LOC_Os09g39300 |
| Deletion | Chr11 | 25043961 | 25044183 | 222 | |
| Deletion | Chr2 | 29835722 | 29835723 | 1 | LOC_Os02g48740 |
| Deletion | Chr3 | 32937915 | 32937916 | 1 | |
| Deletion | Chr1 | 35661897 | 35661903 | 6 |
Insertions: 6
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 1537896 | 1669059 | 2 |
| Inversion | Chr5 | 1538675 | 1693135 | |
| Inversion | Chr11 | 23896286 | 24054035 | LOC_Os11g40350 |
| Inversion | Chr11 | 23896314 | 24054041 | LOC_Os11g40350 |
| Inversion | Chr4 | 35061514 | 35336794 | |
| Inversion | Chr4 | 35061523 | 35336800 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 6001509 | Chr2 | 16964432 |
