Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3263-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3263-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 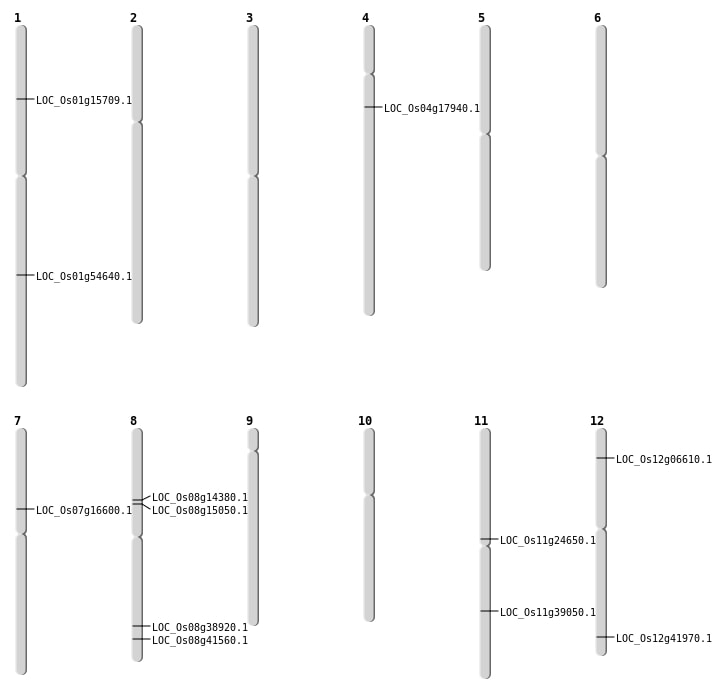
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10985756 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 22749028 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 32080844 | G-A | HET | INTRON | |
| SBS | Chr1 | 33323751 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 33323762 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 39798664 | T-A | HET | START_GAINED | |
| SBS | Chr1 | 42622671 | A-C | HET | INTRON | |
| SBS | Chr10 | 6371448 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 11270396 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 14061845 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g24650 |
| SBS | Chr11 | 15932895 | A-C | HET | INTRON | |
| SBS | Chr11 | 19097742 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 23816458 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 13507526 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22032871 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3144309 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 13391062 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 31244506 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 35019725 | T-A | HET | INTRON | |
| SBS | Chr3 | 35019728 | T-G | HET | INTRON | |
| SBS | Chr4 | 1737413 | G-A | HET | INTRON | |
| SBS | Chr5 | 28575974 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 21533960 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 105686 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 20866230 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2269115 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7874066 | C-G | HET | INTRON | |
| SBS | Chr7 | 9739847 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16600 |
| SBS | Chr8 | 22052305 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 24598577 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g38920 |
| SBS | Chr8 | 26421410 | C-T | HET | INTRON | |
| SBS | Chr8 | 26737365 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 8611477 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 15229798 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8306272 | C-A | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2636899 | 2636900 | 1 | |
| Deletion | Chr12 | 3202022 | 3202034 | 12 | LOC_Os12g06610 |
| Deletion | Chr7 | 4539115 | 4539117 | 2 | |
| Deletion | Chr6 | 4954847 | 4954849 | 2 | |
| Deletion | Chr3 | 5330457 | 5330458 | 1 | |
| Deletion | Chr10 | 5491082 | 5491083 | 1 | |
| Deletion | Chr11 | 8275507 | 8275512 | 5 | |
| Deletion | Chr8 | 8625693 | 8625702 | 9 | LOC_Os08g14380 |
| Deletion | Chr1 | 8836230 | 8836264 | 34 | LOC_Os01g15709 |
| Deletion | Chr8 | 9099344 | 9099345 | 1 | LOC_Os08g15050 |
| Deletion | Chr4 | 9865346 | 9865348 | 2 | LOC_Os04g17940 |
| Deletion | Chr4 | 13656803 | 13656808 | 5 | |
| Deletion | Chr4 | 15098265 | 15098266 | 1 | |
| Deletion | Chr7 | 16287285 | 16287289 | 4 | |
| Deletion | Chr2 | 16948324 | 16948328 | 4 | |
| Deletion | Chr8 | 17868834 | 17868836 | 2 | |
| Deletion | Chr4 | 18526251 | 18526252 | 1 | |
| Deletion | Chr3 | 20213756 | 20213757 | 1 | |
| Deletion | Chr6 | 21268955 | 21268956 | 1 | |
| Deletion | Chr1 | 22749637 | 22749638 | 1 | |
| Deletion | Chr11 | 23250292 | 23250303 | 11 | LOC_Os11g39050 |
| Deletion | Chr12 | 26026839 | 26026841 | 2 | LOC_Os12g41970 |
| Deletion | Chr8 | 26255832 | 26255844 | 12 | LOC_Os08g41560 |
| Deletion | Chr8 | 27181506 | 27181517 | 11 | |
| Deletion | Chr7 | 28263198 | 28263200 | 2 | |
| Deletion | Chr5 | 29873966 | 29873967 | 1 | |
| Deletion | Chr1 | 32934332 | 32934333 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 20763484 | 20763484 | 1 | |
| Insertion | Chr4 | 28151654 | 28151655 | 2 | |
| Insertion | Chr5 | 20704701 | 20704722 | 22 | |
| Insertion | Chr7 | 4279975 | 4279976 | 2 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 7765123 | 7944366 | |
| Inversion | Chr9 | 7765133 | 7944366 | |
| Inversion | Chr1 | 30929517 | 31435102 | LOC_Os01g54640 |
| Inversion | Chr1 | 30929528 | 31761523 | |
| Inversion | Chr1 | 31327182 | 31761514 | |
| Inversion | Chr1 | 31327188 | 31327739 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 6311480 | Chr7 | 25640371 | |
| Translocation | Chr8 | 6428781 | Chr7 | 25640365 |
