Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3278-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3278-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 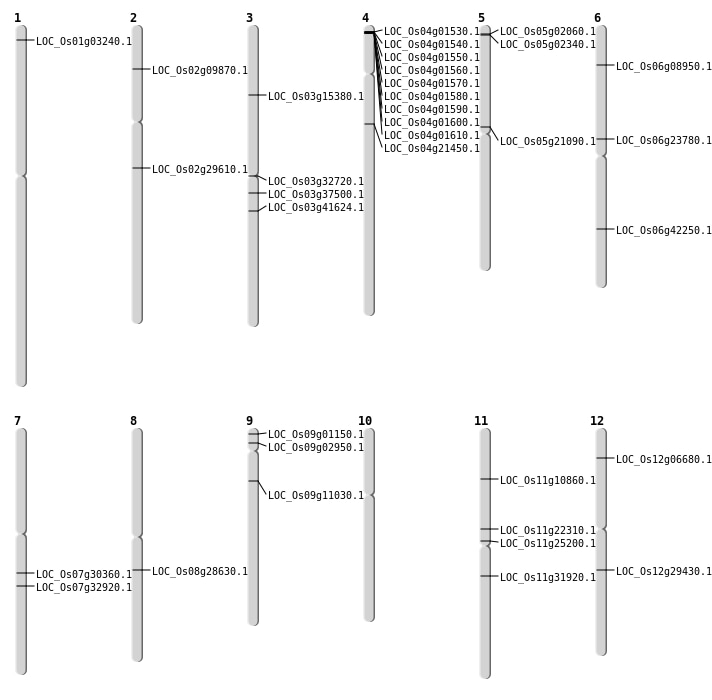
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12082437 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 12082438 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 29877930 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 12788717 | G-A | HOMO | STOP_GAINED | LOC_Os11g22310 |
| SBS | Chr11 | 8180714 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 10176800 | G-A | HET | INTRON | |
| SBS | Chr12 | 1211586 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 22568465 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 2341745 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 13615633 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 21073669 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 11194435 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 29311751 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2580464 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 12398655 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g21090 |
| SBS | Chr5 | 1722423 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 789896 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13338408 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 16448627 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 26132496 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 4204105 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6572762 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 14391753 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr7 | 19684105 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g32920 |
| SBS | Chr7 | 21916099 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 16959349 | G-T | HET | INTRON | |
| SBS | Chr9 | 18172735 | G-A | HET | INTERGENIC |
Deletions: 66
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 360001 | 414000 | 53999 | 10 |
| Deletion | Chr9 | 639736 | 639737 | 1 | |
| Deletion | Chr11 | 728595 | 728604 | 9 | |
| Deletion | Chr1 | 1313385 | 1313386 | 1 | LOC_Os01g03240 |
| Deletion | Chr9 | 1383418 | 1383419 | 1 | LOC_Os09g02950 |
| Deletion | Chr10 | 1478545 | 1478547 | 2 | |
| Deletion | Chr10 | 3390729 | 3390751 | 22 | |
| Deletion | Chr3 | 4295001 | 4310000 | 14999 | LOC_Os03g32720 |
| Deletion | Chr9 | 4753131 | 4753204 | 73 | |
| Deletion | Chr9 | 4803131 | 4803132 | 1 | |
| Deletion | Chr2 | 5096851 | 5096852 | 1 | LOC_Os02g09870 |
| Deletion | Chr12 | 5370558 | 5370559 | 1 | |
| Deletion | Chr5 | 5540550 | 5540568 | 18 | |
| Deletion | Chr11 | 5831539 | 5831540 | 1 | |
| Deletion | Chr11 | 5982724 | 5982727 | 3 | LOC_Os11g10860 |
| Deletion | Chr5 | 6310337 | 6310387 | 50 | |
| Deletion | Chr10 | 6326382 | 6326383 | 1 | |
| Deletion | Chr5 | 6411381 | 6411382 | 1 | |
| Deletion | Chr4 | 6726292 | 6726293 | 1 | |
| Deletion | Chr6 | 7979725 | 7979736 | 11 | |
| Deletion | Chr6 | 8460545 | 8460546 | 1 | |
| Deletion | Chr12 | 8522689 | 8522692 | 3 | |
| Deletion | Chr7 | 9236040 | 9236041 | 1 | |
| Deletion | Chr12 | 9705869 | 9705870 | 1 | |
| Deletion | Chr11 | 9981924 | 9981925 | 1 | |
| Deletion | Chr1 | 11489604 | 11489605 | 1 | |
| Deletion | Chr11 | 11501358 | 11501359 | 1 | |
| Deletion | Chr9 | 11645554 | 11645624 | 70 | |
| Deletion | Chr8 | 11825151 | 11825218 | 67 | |
| Deletion | Chr12 | 11911824 | 11911825 | 1 | |
| Deletion | Chr5 | 12019469 | 12019470 | 1 | |
| Deletion | Chr1 | 12116038 | 12116039 | 1 | |
| Deletion | Chr7 | 12164891 | 12164893 | 2 | |
| Deletion | Chr7 | 12240311 | 12240312 | 1 | |
| Deletion | Chr1 | 12844290 | 12844291 | 1 | |
| Deletion | Chr5 | 12954557 | 12954558 | 1 | |
| Deletion | Chr8 | 13015583 | 13015584 | 1 | |
| Deletion | Chr9 | 13750392 | 13750393 | 1 | |
| Deletion | Chr10 | 14078514 | 14078515 | 1 | |
| Deletion | Chr10 | 14212029 | 14212030 | 1 | |
| Deletion | Chr4 | 14263621 | 14263622 | 1 | |
| Deletion | Chr11 | 14358048 | 14358049 | 1 | LOC_Os11g25200 |
| Deletion | Chr11 | 14358365 | 14358366 | 1 | |
| Deletion | Chr8 | 14905489 | 14905490 | 1 | |
| Deletion | Chr7 | 15827465 | 15827510 | 45 | |
| Deletion | Chr2 | 16564181 | 16564278 | 97 | |
| Deletion | Chr11 | 16809509 | 16809572 | 63 | |
| Deletion | Chr12 | 17479698 | 17479748 | 50 | LOC_Os12g29430 |
| Deletion | Chr7 | 20498435 | 20498436 | 1 | |
| Deletion | Chr3 | 20810052 | 20810053 | 1 | LOC_Os03g37500 |
| Deletion | Chr6 | 21771389 | 21771452 | 63 | |
| Deletion | Chr4 | 22002195 | 22002196 | 1 | |
| Deletion | Chr6 | 22384700 | 22384764 | 64 | |
| Deletion | Chr5 | 23052307 | 23052308 | 1 | |
| Deletion | Chr11 | 24645115 | 24645116 | 1 | |
| Deletion | Chr4 | 24733286 | 24733293 | 7 | |
| Deletion | Chr6 | 25371892 | 25371893 | 1 | LOC_Os06g42250 |
| Deletion | Chr11 | 28628162 | 28628163 | 1 | |
| Deletion | Chr4 | 29713232 | 29713236 | 4 | |
| Deletion | Chr1 | 29866893 | 29866895 | 2 | |
| Deletion | Chr2 | 30793140 | 30793141 | 1 | |
| Deletion | Chr1 | 31339678 | 31339679 | 1 | |
| Deletion | Chr4 | 33293328 | 33293370 | 42 | |
| Deletion | Chr1 | 34069850 | 34069853 | 3 | |
| Deletion | Chr3 | 35686126 | 35686127 | 1 | |
| Deletion | Chr1 | 42526281 | 42526282 | 1 |
Insertions: 51
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 20181135 | 20181137 | 3 | |
| Insertion | Chr1 | 22508981 | 22508981 | 1 | |
| Insertion | Chr1 | 28135120 | 28135127 | 8 | |
| Insertion | Chr1 | 4555888 | 4555888 | 1 | |
| Insertion | Chr10 | 10067093 | 10067093 | 1 | |
| Insertion | Chr10 | 10455541 | 10455541 | 1 | |
| Insertion | Chr10 | 164450 | 164450 | 1 | |
| Insertion | Chr10 | 3923863 | 3923863 | 1 | |
| Insertion | Chr10 | 5665704 | 5665706 | 3 | |
| Insertion | Chr10 | 8750831 | 8750831 | 1 | |
| Insertion | Chr10 | 8854365 | 8854365 | 1 | |
| Insertion | Chr11 | 12649782 | 12649782 | 1 | |
| Insertion | Chr11 | 18074281 | 18074282 | 2 | |
| Insertion | Chr11 | 18759708 | 18759711 | 4 | LOC_Os11g31920 |
| Insertion | Chr11 | 21226713 | 21226713 | 1 | |
| Insertion | Chr11 | 23031529 | 23031550 | 22 | |
| Insertion | Chr11 | 7021038 | 7021038 | 1 | |
| Insertion | Chr12 | 14610999 | 14611000 | 2 | |
| Insertion | Chr2 | 11030182 | 11030192 | 11 | |
| Insertion | Chr2 | 24766580 | 24766580 | 1 | |
| Insertion | Chr2 | 33687580 | 33687580 | 1 | |
| Insertion | Chr3 | 13514048 | 13514048 | 1 | |
| Insertion | Chr3 | 23143532 | 23143532 | 1 | LOC_Os03g41624 |
| Insertion | Chr3 | 25259054 | 25259057 | 4 | |
| Insertion | Chr3 | 31280028 | 31280037 | 10 | |
| Insertion | Chr3 | 8245080 | 8245134 | 55 | |
| Insertion | Chr4 | 22528362 | 22528406 | 45 | |
| Insertion | Chr4 | 32269364 | 32269364 | 1 | |
| Insertion | Chr4 | 4268572 | 4268572 | 1 | |
| Insertion | Chr4 | 7871912 | 7871934 | 23 | |
| Insertion | Chr5 | 10211480 | 10211480 | 1 | |
| Insertion | Chr5 | 5661449 | 5661449 | 1 | |
| Insertion | Chr6 | 11372200 | 11372201 | 2 | |
| Insertion | Chr6 | 16436960 | 16436998 | 39 | |
| Insertion | Chr6 | 29316479 | 29316479 | 1 | |
| Insertion | Chr6 | 4514099 | 4514099 | 1 | LOC_Os06g08950 |
| Insertion | Chr6 | 7028658 | 7028729 | 72 | |
| Insertion | Chr7 | 13114638 | 13114638 | 1 | |
| Insertion | Chr7 | 15761476 | 15761477 | 2 | |
| Insertion | Chr7 | 15967538 | 15967538 | 1 | |
| Insertion | Chr7 | 17430499 | 17430501 | 3 | |
| Insertion | Chr7 | 17968057 | 17968057 | 1 | LOC_Os07g30360 |
| Insertion | Chr7 | 29283370 | 29283370 | 1 | |
| Insertion | Chr8 | 12369352 | 12369352 | 1 | |
| Insertion | Chr8 | 21275090 | 21275160 | 71 | |
| Insertion | Chr8 | 22240896 | 22240896 | 1 | |
| Insertion | Chr9 | 157564 | 157567 | 4 | LOC_Os09g01150 |
| Insertion | Chr9 | 5365238 | 5365238 | 1 | |
| Insertion | Chr9 | 5740439 | 5740439 | 1 | |
| Insertion | Chr9 | 8227266 | 8227268 | 3 | |
| Insertion | Chr9 | 9259289 | 9259289 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 600466 | 696973 | LOC_Os05g02060 |
Translocations: 18
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 600434 | Chr4 | 4408500 | LOC_Os05g02060 |
| Translocation | Chr5 | 696967 | Chr4 | 4408498 | |
| Translocation | Chr4 | 724907 | Chr3 | 31273242 | |
| Translocation | Chr12 | 3253647 | Chr8 | 17492515 | 2 |
| Translocation | Chr9 | 6127699 | Chr5 | 754968 | 2 |
| Translocation | Chr9 | 6385471 | Chr1 | 10914859 | |
| Translocation | Chr5 | 8562078 | Chr1 | 40306295 | |
| Translocation | Chr8 | 8711799 | Chr3 | 8414052 | LOC_Os03g15380 |
| Translocation | Chr9 | 10396957 | Chr1 | 42148951 | |
| Translocation | Chr10 | 13764672 | Chr2 | 29617583 | |
| Translocation | Chr6 | 13897631 | Chr1 | 8742073 | LOC_Os06g23780 |
| Translocation | Chr6 | 13898931 | Chr1 | 8742070 | |
| Translocation | Chr5 | 15019882 | Chr2 | 17615209 | LOC_Os02g29610 |
| Translocation | Chr8 | 15027895 | Chr7 | 11295924 | |
| Translocation | Chr6 | 15873932 | Chr1 | 8112024 | |
| Translocation | Chr12 | 20230686 | Chr2 | 24154265 | |
| Translocation | Chr4 | 22148736 | Chr3 | 15326889 | |
| Translocation | Chr11 | 23663020 | Chr6 | 11605812 |
