Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3287-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3287-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 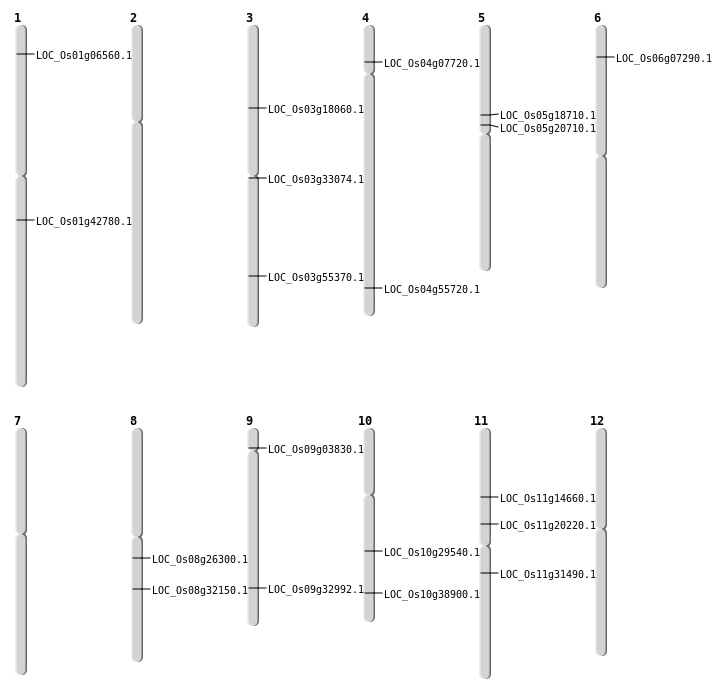
|
| Variant Information |
|---|
Single base substitutions: 61
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10058378 | T-A | HET | ||
| SBS | Chr1 | 11763896 | C-T | HET | ||
| SBS | Chr1 | 13212912 | G-A | HET | ||
| SBS | Chr1 | 24344877 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g42780 |
| SBS | Chr1 | 24583616 | G-A | Homo | ||
| SBS | Chr10 | 20198640 | A-T | HET | ||
| SBS | Chr10 | 2147823 | A-G | Homo | ||
| SBS | Chr10 | 5218310 | T-A | HET | ||
| SBS | Chr10 | 9940966 | A-T | HET | ||
| SBS | Chr11 | 11677785 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g20220 |
| SBS | Chr11 | 18402057 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31490 |
| SBS | Chr11 | 20768048 | C-T | HET | ||
| SBS | Chr11 | 8264015 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g14660 |
| SBS | Chr11 | 835721 | A-G | HET | ||
| SBS | Chr12 | 20343152 | G-C | HET | ||
| SBS | Chr12 | 21057414 | C-T | HET | ||
| SBS | Chr12 | 9113351 | G-T | HET | ||
| SBS | Chr2 | 14386191 | T-C | HET | ||
| SBS | Chr2 | 20098131 | C-T | HET | ||
| SBS | Chr2 | 21978201 | G-A | Homo | ||
| SBS | Chr2 | 22226097 | C-T | HET | ||
| SBS | Chr2 | 2482288 | T-C | HET | ||
| SBS | Chr2 | 26991356 | G-T | Homo | ||
| SBS | Chr2 | 4821227 | A-C | Homo | ||
| SBS | Chr3 | 10068151 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g18060 |
| SBS | Chr3 | 18925828 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33074 |
| SBS | Chr3 | 26459391 | G-A | HET | ||
| SBS | Chr3 | 8449671 | G-A | Homo | ||
| SBS | Chr4 | 10813104 | G-C | Homo | ||
| SBS | Chr4 | 17662219 | C-G | HET | ||
| SBS | Chr4 | 28456327 | A-C | HET | ||
| SBS | Chr4 | 33169382 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g55720 |
| SBS | Chr4 | 6825424 | A-T | HET | ||
| SBS | Chr5 | 10835330 | G-A | Homo | SPLICE_SITE_DONOR | LOC_Os05g18710 |
| SBS | Chr5 | 12140692 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g20710 |
| SBS | Chr5 | 12577402 | A-G | HET | ||
| SBS | Chr5 | 16895478 | C-T | HET | ||
| SBS | Chr5 | 18945041 | C-A | HET | ||
| SBS | Chr5 | 26333603 | T-C | HET | ||
| SBS | Chr5 | 28398560 | T-A | HET | ||
| SBS | Chr6 | 13261018 | G-A | HET | ||
| SBS | Chr6 | 13287525 | C-G | HET | ||
| SBS | Chr6 | 19750704 | C-T | HET | ||
| SBS | Chr6 | 3498811 | G-T | HET | SPLICE_SITE_DONOR | LOC_Os06g07290 |
| SBS | Chr6 | 4986931 | A-C | HET | ||
| SBS | Chr6 | 5544759 | A-T | HET | ||
| SBS | Chr6 | 8615563 | T-G | Homo | ||
| SBS | Chr7 | 26648928 | G-A | HET | ||
| SBS | Chr7 | 27155914 | G-C | HET | ||
| SBS | Chr7 | 29682658 | G-T | HET | ||
| SBS | Chr8 | 11368886 | G-A | HET | ||
| SBS | Chr8 | 16041212 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g26300 |
| SBS | Chr8 | 16793210 | G-A | HET | ||
| SBS | Chr8 | 17095192 | G-A | HET | ||
| SBS | Chr8 | 19946813 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g32150 |
| SBS | Chr9 | 12766382 | C-T | HET | ||
| SBS | Chr9 | 16361710 | C-A | HET | ||
| SBS | Chr9 | 17975203 | C-T | HET | ||
| SBS | Chr9 | 2178080 | T-G | HET | ||
| SBS | Chr9 | 2521941 | C-T | HET | ||
| SBS | Chr9 | 8634933 | T-C | HET |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1943295 | 1943296 | 1 | LOC_Os09g03830 |
| Deletion | Chr5 | 2049550 | 2049556 | 6 | |
| Deletion | Chr5 | 2822351 | 2822352 | 1 | |
| Deletion | Chr12 | 8797140 | 8797142 | 2 | |
| Deletion | Chr12 | 9113366 | 9113368 | 2 | |
| Deletion | Chr4 | 10449644 | 10449646 | 2 | |
| Deletion | Chr7 | 10708826 | 10708835 | 9 | |
| Deletion | Chr11 | 10847456 | 10847457 | 1 | |
| Deletion | Chr3 | 11018353 | 11018359 | 6 | |
| Deletion | Chr10 | 11487628 | 11487629 | 1 | |
| Deletion | Chr5 | 12196140 | 12196158 | 18 | |
| Deletion | Chr3 | 13360650 | 13360661 | 11 | |
| Deletion | Chr8 | 17155496 | 17155497 | 1 | |
| Deletion | Chr12 | 19116566 | 19116567 | 1 | |
| Deletion | Chr6 | 19660768 | 19660770 | 2 | |
| Deletion | Chr3 | 20672833 | 20672841 | 8 | |
| Deletion | Chr10 | 20728018 | 20728019 | 1 | LOC_Os10g38900 |
| Deletion | Chr10 | 22649903 | 22649905 | 2 | |
| Deletion | Chr6 | 25586820 | 25586873 | 53 | |
| Deletion | Chr8 | 25735484 | 25735485 | 1 | |
| Deletion | Chr6 | 26213878 | 26213885 | 7 | |
| Deletion | Chr3 | 28507842 | 28507843 | 1 | |
| Deletion | Chr3 | 31434908 | 31434911 | 3 | |
| Deletion | Chr2 | 32661720 | 32661731 | 11 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 28683435 | 28683437 | 3 | |
| Insertion | Chr4 | 27444221 | 27444223 | 3 | |
| Insertion | Chr5 | 20220797 | 20220797 | 1 | |
| Insertion | Chr5 | 20323963 | 20323968 | 6 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 8966855 | 9086946 | |
| Inversion | Chr9 | 8966866 | 9086954 | |
| Inversion | Chr10 | 15348936 | 15349389 | LOC_Os10g29540 |
| Inversion | Chr12 | 17972533 | 18616773 | |
| Inversion | Chr12 | 17972542 | 18616778 | |
| Inversion | Chr1 | 22905292 | 22905530 | |
| Inversion | Chr2 | 28103743 | 28104000 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 10156938 | Chr1 | 3091823 | 2 |
| Translocation | Chr9 | 17271819 | Chr4 | 4107386 | 2 |
| Translocation | Chr9 | 18246920 | Chr3 | 31502590 | |
| Translocation | Chr9 | 18246928 | Chr3 | 31502032 | 2 |
| Translocation | Chr12 | 24236938 | Chr9 | 19697761 | 2 |
