Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN337-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN337-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 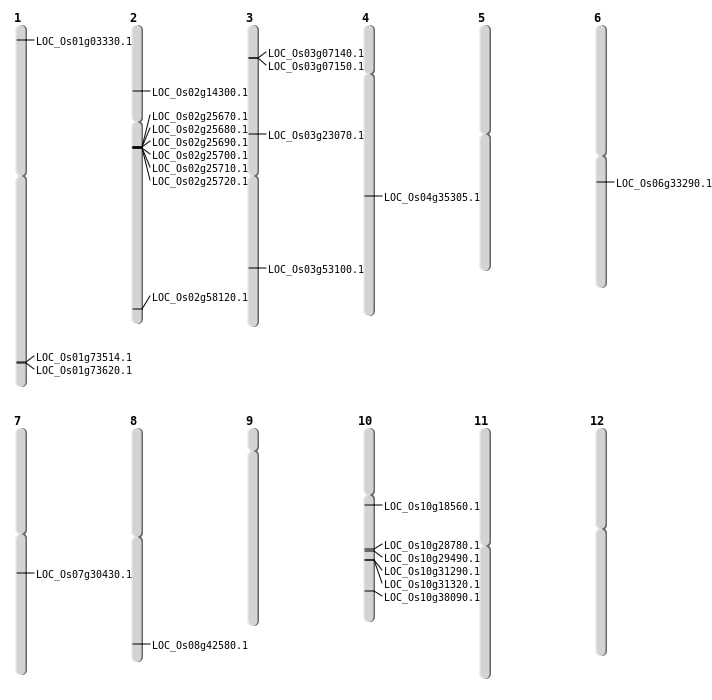
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 37002457 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 15761259 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 1895170 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 13032440 | G-A | HET | INTRON | |
| SBS | Chr11 | 19714826 | T-A | HET | INTRON | |
| SBS | Chr12 | 1420273 | T-C | HOMO | INTRON | |
| SBS | Chr12 | 1789720 | C-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 1789721 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 312314 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 20101805 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 29857595 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 30326480 | A-G | HET | INTRON | |
| SBS | Chr2 | 35569684 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 6494962 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 2367273 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 26108759 | G-C | HET | INTRON | |
| SBS | Chr3 | 9470125 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 20323978 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21470500 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g35305 |
| SBS | Chr5 | 4794471 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 24180715 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 30960943 | C-A | HET | INTRON | |
| SBS | Chr6 | 4058935 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 13044831 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 18013017 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30430 |
| SBS | Chr7 | 18162574 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 99096 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 25054498 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 4892650 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 4892651 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 7516616 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 17107903 | G-A | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1344807 | 1344810 | 3 | LOC_Os01g03330 |
| Deletion | Chr3 | 3657338 | 3657340 | 2 | 2 |
| Deletion | Chr12 | 5055854 | 5055855 | 1 | |
| Deletion | Chr2 | 7852218 | 7852228 | 10 | LOC_Os02g14300 |
| Deletion | Chr10 | 8799769 | 8799783 | 14 | |
| Deletion | Chr10 | 9261875 | 9261905 | 30 | |
| Deletion | Chr3 | 13336901 | 13336918 | 17 | LOC_Os03g23070 |
| Deletion | Chr2 | 15013001 | 15062000 | 49000 | 6 |
| Deletion | Chr10 | 16415275 | 16415277 | 2 | LOC_Os10g31290 |
| Deletion | Chr10 | 16426808 | 16426841 | 33 | LOC_Os10g31320 |
| Deletion | Chr12 | 17984160 | 17984161 | 1 | |
| Deletion | Chr11 | 18778234 | 18778257 | 23 | |
| Deletion | Chr6 | 19381144 | 19381154 | 10 | LOC_Os06g33290 |
| Deletion | Chr1 | 19567187 | 19567190 | 3 | |
| Deletion | Chr10 | 20395129 | 20395130 | 1 | LOC_Os10g38090 |
| Deletion | Chr2 | 21685872 | 21685897 | 25 | |
| Deletion | Chr8 | 26911883 | 26911886 | 3 | LOC_Os08g42580 |
| Deletion | Chr3 | 28118313 | 28118316 | 3 | |
| Deletion | Chr11 | 28790449 | 28790453 | 4 | |
| Deletion | Chr3 | 30452767 | 30452768 | 1 | LOC_Os03g53100 |
| Deletion | Chr2 | 35569768 | 35569777 | 9 | LOC_Os02g58120 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 9413777 | 9413778 | 2 | LOC_Os10g18560 |
| Insertion | Chr11 | 22223380 | 22223383 | 4 | |
| Insertion | Chr2 | 19427799 | 19427804 | 6 | |
| Insertion | Chr5 | 2389364 | 2389364 | 1 | |
| Insertion | Chr6 | 10367526 | 10367526 | 1 | |
| Insertion | Chr7 | 24289012 | 24289013 | 2 | |
| Insertion | Chr8 | 9212042 | 9212042 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 15005236 | 15317177 | LOC_Os10g29490 |
| Inversion | Chr10 | 15011757 | 15317176 | 2 |
| Inversion | Chr1 | 42595960 | 42647554 | 2 |
No Translocation
