Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3440-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3440-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 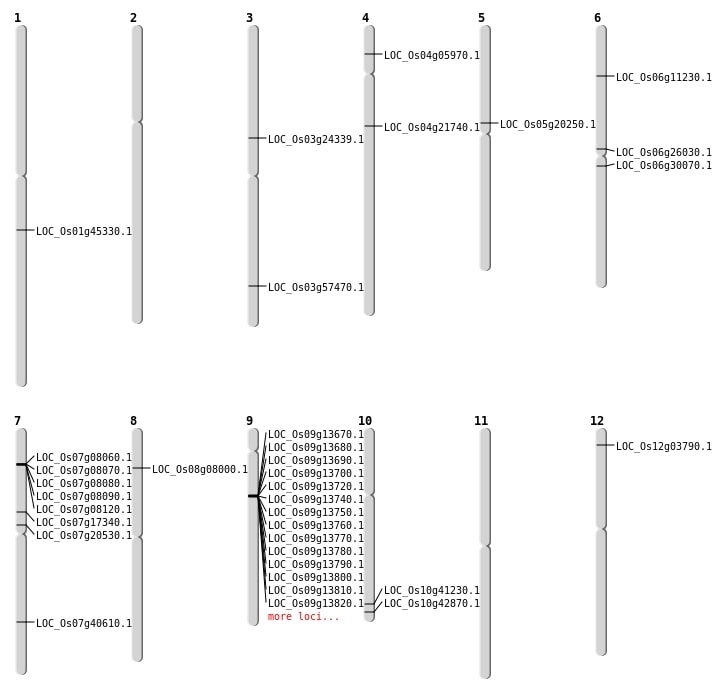
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23677017 | G-T | HET | ||
| SBS | Chr1 | 25732310 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45330 |
| SBS | Chr1 | 35948615 | T-A | HET | ||
| SBS | Chr10 | 17817440 | G-C | HET | ||
| SBS | Chr10 | 19750295 | C-A | HET | ||
| SBS | Chr10 | 19750296 | C-G | HET | ||
| SBS | Chr10 | 9009645 | G-T | HET | ||
| SBS | Chr11 | 22359590 | G-A | HET | ||
| SBS | Chr11 | 23030924 | C-T | HET | ||
| SBS | Chr11 | 3157220 | T-A | Homo | ||
| SBS | Chr11 | 638208 | G-A | Homo | ||
| SBS | Chr12 | 22963573 | G-T | HET | ||
| SBS | Chr12 | 687737 | G-A | Homo | ||
| SBS | Chr2 | 1274534 | A-T | HET | ||
| SBS | Chr2 | 26848277 | C-T | HET | ||
| SBS | Chr3 | 12476343 | A-G | HET | ||
| SBS | Chr3 | 13549787 | T-A | Homo | ||
| SBS | Chr3 | 13549788 | G-A | Homo | ||
| SBS | Chr3 | 17688293 | A-G | HET | ||
| SBS | Chr3 | 32766071 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g57470 |
| SBS | Chr3 | 5424612 | T-C | HET | ||
| SBS | Chr4 | 10804501 | C-A | Homo | ||
| SBS | Chr4 | 12301006 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g21740 |
| SBS | Chr4 | 18329693 | T-G | Homo | ||
| SBS | Chr4 | 20382711 | G-A | HET | ||
| SBS | Chr4 | 24686294 | T-C | HET | ||
| SBS | Chr4 | 3629176 | G-T | HET | ||
| SBS | Chr4 | 8032979 | C-G | HET | ||
| SBS | Chr5 | 11860525 | C-T | HET | STOP_GAINED | LOC_Os05g20250 |
| SBS | Chr5 | 11897357 | G-T | HET | ||
| SBS | Chr5 | 15003882 | T-C | HET | ||
| SBS | Chr6 | 14790062 | C-T | HET | ||
| SBS | Chr6 | 16686816 | G-T | HET | ||
| SBS | Chr6 | 31041864 | T-A | Homo | ||
| SBS | Chr6 | 4484209 | A-G | HET | ||
| SBS | Chr7 | 10238688 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g17340 |
| SBS | Chr7 | 11895233 | C-T | HET | ||
| SBS | Chr8 | 18815503 | C-T | HET | ||
| SBS | Chr8 | 3163209 | C-T | HET | ||
| SBS | Chr8 | 4525136 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08000 |
| SBS | Chr9 | 20842347 | G-A | Homo |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 687680 | 687714 | 34 | |
| Deletion | Chr6 | 2493298 | 2493301 | 3 | |
| Deletion | Chr4 | 3092669 | 3092675 | 6 | LOC_Os04g05970 |
| Deletion | Chr7 | 4088001 | 4125000 | 36999 | 5 |
| Deletion | Chr11 | 5621882 | 5621887 | 5 | |
| Deletion | Chr6 | 6493824 | 6493826 | 2 | |
| Deletion | Chr9 | 7990001 | 8019000 | 28999 | 4 |
| Deletion | Chr9 | 8028001 | 8055000 | 26999 | 3 |
| Deletion | Chr9 | 8064001 | 8358000 | 293999 | 32 |
| Deletion | Chr9 | 8578001 | 8766000 | 187999 | 25 |
| Deletion | Chr9 | 8774001 | 8797000 | 22999 | 3 |
| Deletion | Chr9 | 8829001 | 8837000 | 7999 | |
| Deletion | Chr9 | 8849001 | 8926000 | 76999 | 8 |
| Deletion | Chr9 | 8929001 | 9018000 | 88999 | 7 |
| Deletion | Chr9 | 9260001 | 9298000 | 37999 | 2 |
| Deletion | Chr9 | 9911935 | 9911941 | 6 | LOC_Os09g16240 |
| Deletion | Chr9 | 12912526 | 12912527 | 1 | |
| Deletion | Chr3 | 13843416 | 13843417 | 1 | LOC_Os03g24339 |
| Deletion | Chr6 | 15238670 | 15238690 | 20 | LOC_Os06g26030 |
| Deletion | Chr5 | 16675391 | 16675398 | 7 | |
| Deletion | Chr9 | 16887955 | 16887956 | 1 | |
| Deletion | Chr11 | 17234357 | 17234358 | 1 | |
| Deletion | Chr6 | 17329547 | 17329548 | 1 | LOC_Os06g30070 |
| Deletion | Chr7 | 21785156 | 21785157 | 1 | |
| Deletion | Chr1 | 21800739 | 21800758 | 19 | |
| Deletion | Chr1 | 21928522 | 21928528 | 6 | |
| Deletion | Chr10 | 22153748 | 22153750 | 2 | LOC_Os10g41230 |
| Deletion | Chr4 | 23082871 | 23082872 | 1 | |
| Deletion | Chr2 | 30585147 | 30585149 | 2 | |
| Deletion | Chr3 | 33293936 | 33293937 | 1 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 10449345 | 10449346 | 2 | |
| Insertion | Chr10 | 23114769 | 23114771 | 3 | LOC_Os10g42870 |
| Insertion | Chr12 | 22564309 | 22564309 | 1 | |
| Insertion | Chr2 | 33454857 | 33454857 | 1 | |
| Insertion | Chr6 | 15447616 | 15447617 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 1542987 | 1551591 | LOC_Os12g03790 |
| Inversion | Chr7 | 6549996 | 6985730 | |
| Inversion | Chr8 | 14534925 | 14548971 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1542996 | Chr4 | 516800 | LOC_Os12g03790 |
| Translocation | Chr3 | 5096470 | Chr1 | 27526753 | |
| Translocation | Chr9 | 8843428 | Chr7 | 24331917 | 2 |
| Translocation | Chr7 | 11859814 | Chr6 | 2921274 | LOC_Os07g20530 |
| Translocation | Chr11 | 19748564 | Chr6 | 5893165 | 2 |
