Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3667-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3667-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 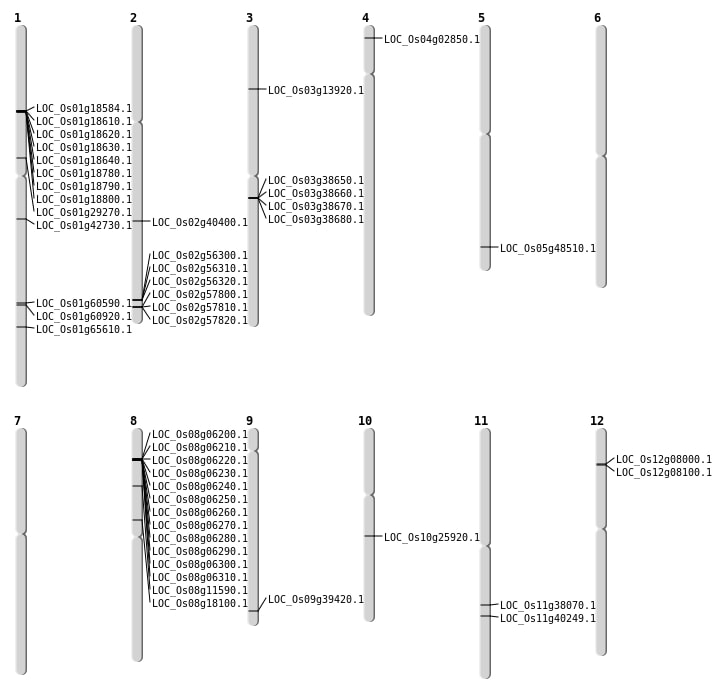
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15012401 | T-C | HET | ||
| SBS | Chr1 | 16412683 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g29270 |
| SBS | Chr1 | 25930480 | T-A | HET | ||
| SBS | Chr1 | 338490 | A-G | Homo | ||
| SBS | Chr1 | 35025934 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g60590 |
| SBS | Chr1 | 35239155 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g60920 |
| SBS | Chr1 | 37062040 | C-T | HET | ||
| SBS | Chr1 | 41705787 | C-A | HET | ||
| SBS | Chr1 | 41858040 | T-A | HET | ||
| SBS | Chr10 | 12656007 | G-A | HET | ||
| SBS | Chr10 | 5003382 | C-A | HET | ||
| SBS | Chr11 | 10474882 | C-A | HET | ||
| SBS | Chr11 | 9423338 | A-T | HET | ||
| SBS | Chr12 | 19170667 | C-T | HET | ||
| SBS | Chr12 | 4066622 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g08000 |
| SBS | Chr12 | 9373698 | G-A | HET | ||
| SBS | Chr2 | 16017955 | G-A | HET | ||
| SBS | Chr2 | 31717231 | A-T | HET | ||
| SBS | Chr2 | 35631097 | C-T | HET | ||
| SBS | Chr2 | 4849583 | C-T | HET | ||
| SBS | Chr3 | 11830220 | T-G | HET | ||
| SBS | Chr3 | 16058877 | A-G | HET | ||
| SBS | Chr3 | 16217767 | C-T | Homo | ||
| SBS | Chr3 | 29119072 | A-G | HET | ||
| SBS | Chr3 | 7378880 | G-T | HET | ||
| SBS | Chr3 | 8978205 | G-A | Homo | ||
| SBS | Chr4 | 8863079 | T-C | HET | ||
| SBS | Chr4 | 9238640 | G-A | HET | ||
| SBS | Chr5 | 27804957 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g48510 |
| SBS | Chr6 | 23910889 | C-T | HET | ||
| SBS | Chr6 | 26890220 | C-G | HET | ||
| SBS | Chr6 | 8123707 | C-G | HET | ||
| SBS | Chr7 | 19065969 | A-G | HET | ||
| SBS | Chr7 | 24810559 | T-A | Homo | ||
| SBS | Chr7 | 381070 | A-C | Homo | ||
| SBS | Chr8 | 11096211 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g18100 |
| SBS | Chr8 | 11096232 | A-T | HET | ||
| SBS | Chr8 | 19208850 | A-G | Homo | ||
| SBS | Chr8 | 4365469 | C-T | HET | ||
| SBS | Chr8 | 6359561 | G-C | Homo | ||
| SBS | Chr8 | 6964878 | G-C | HET | ||
| SBS | Chr9 | 13779324 | A-G | HET | ||
| SBS | Chr9 | 16609127 | C-A | Homo | ||
| SBS | Chr9 | 22670836 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g39420 |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 61680 | 61707 | 27 | |
| Deletion | Chr8 | 3420001 | 3479000 | 58999 | 12 |
| Deletion | Chr9 | 4337283 | 4337295 | 12 | |
| Deletion | Chr2 | 5293630 | 5293631 | 1 | |
| Deletion | Chr4 | 5671722 | 5671723 | 1 | |
| Deletion | Chr1 | 6723579 | 6723584 | 5 | |
| Deletion | Chr8 | 6794918 | 6794923 | 5 | LOC_Os08g11590 |
| Deletion | Chr3 | 7553402 | 7553404 | 2 | LOC_Os03g13920 |
| Deletion | Chr5 | 9131834 | 9131842 | 8 | |
| Deletion | Chr2 | 9139505 | 9139653 | 148 | |
| Deletion | Chr5 | 9845494 | 9845500 | 6 | |
| Deletion | Chr1 | 10470001 | 10498000 | 27999 | 5 |
| Deletion | Chr1 | 10578001 | 10628000 | 49999 | 3 |
| Deletion | Chr8 | 16576616 | 16576617 | 1 | |
| Deletion | Chr9 | 16630964 | 16630965 | 1 | |
| Deletion | Chr12 | 16658378 | 16658380 | 2 | |
| Deletion | Chr5 | 17095525 | 17095534 | 9 | |
| Deletion | Chr9 | 18588964 | 18588975 | 11 | |
| Deletion | Chr3 | 21458001 | 21475000 | 16999 | 4 |
| Deletion | Chr5 | 22279666 | 22279667 | 1 | |
| Deletion | Chr3 | 24834596 | 24834599 | 3 | |
| Deletion | Chr2 | 34464001 | 34478000 | 13999 | 3 |
| Deletion | Chr2 | 35393001 | 35418000 | 24999 | 3 |
| Deletion | Chr1 | 39975278 | 39975284 | 6 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 32988340 | 32988343 | 4 | |
| Insertion | Chr11 | 27121659 | 27121866 | 208 | |
| Insertion | Chr12 | 4130499 | 4130499 | 1 | LOC_Os12g08100 |
| Insertion | Chr5 | 3532026 | 3532028 | 3 | |
| Insertion | Chr6 | 13982283 | 13982283 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 27978715 | 28708331 |
Translocations: 13
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 126515 | Chr7 | 14047221 | |
| Translocation | Chr8 | 1361488 | Chr2 | 2418050 | |
| Translocation | Chr5 | 1851293 | Chr4 | 1112574 | LOC_Os04g02850 |
| Translocation | Chr8 | 3074105 | Chr1 | 24215764 | |
| Translocation | Chr8 | 3074196 | Chr1 | 24316812 | LOC_Os01g42730 |
| Translocation | Chr3 | 5504283 | Chr1 | 38091882 | LOC_Os01g65610 |
| Translocation | Chr10 | 5720401 | Chr2 | 24465250 | LOC_Os02g40400 |
| Translocation | Chr4 | 8353881 | Chr1 | 32282138 | |
| Translocation | Chr12 | 9228988 | Chr4 | 7080256 | |
| Translocation | Chr11 | 22579375 | Chr8 | 3761939 | LOC_Os11g38070 |
| Translocation | Chr11 | 24005113 | Chr2 | 8611395 | LOC_Os11g40249 |
| Translocation | Chr11 | 26067191 | Chr10 | 13432175 | LOC_Os10g25920 |
| Translocation | Chr11 | 27311378 | Chr4 | 20036672 |
