Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3714-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3714-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 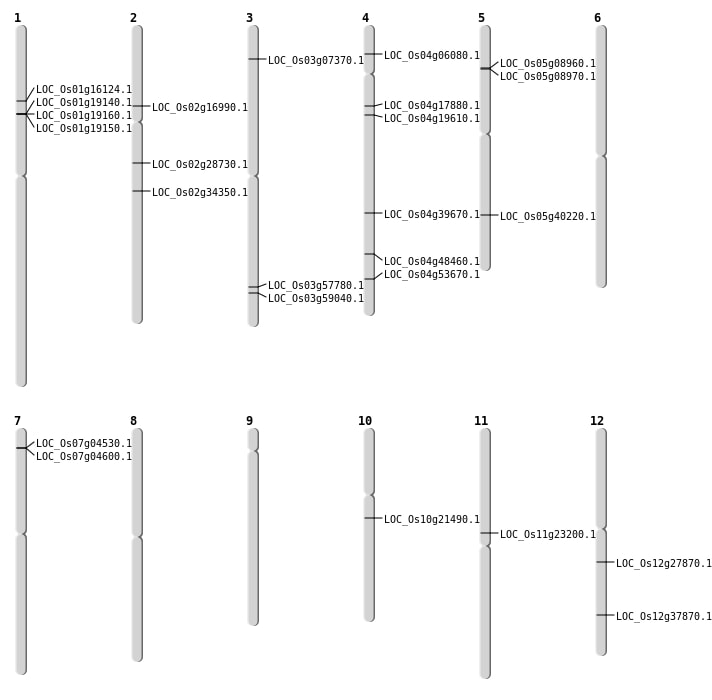
|
| Variant Information |
|---|
Single base substitutions: 56
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10532504 | T-C | HET | ||
| SBS | Chr1 | 15443981 | C-T | HET | ||
| SBS | Chr1 | 17064482 | A-T | HET | ||
| SBS | Chr1 | 26474428 | G-A | HET | ||
| SBS | Chr1 | 27281072 | G-A | HET | ||
| SBS | Chr1 | 29333294 | G-A | HET | ||
| SBS | Chr1 | 39160822 | G-T | Homo | ||
| SBS | Chr1 | 5286007 | T-A | HET | ||
| SBS | Chr1 | 6104311 | G-T | HET | ||
| SBS | Chr10 | 11007907 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g21490 |
| SBS | Chr10 | 12201047 | T-C | HET | ||
| SBS | Chr10 | 21661501 | T-A | HET | ||
| SBS | Chr10 | 21661502 | C-T | HET | ||
| SBS | Chr10 | 21661503 | A-T | HET | ||
| SBS | Chr10 | 824779 | G-A | HET | ||
| SBS | Chr11 | 18978244 | G-A | HET | ||
| SBS | Chr11 | 21354702 | C-T | Homo | ||
| SBS | Chr11 | 21354703 | C-T | Homo | ||
| SBS | Chr12 | 14726685 | T-C | HET | ||
| SBS | Chr12 | 18394749 | C-A | HET | ||
| SBS | Chr12 | 19983708 | G-A | Homo | ||
| SBS | Chr12 | 23275427 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g37870 |
| SBS | Chr12 | 8947468 | G-A | HET | ||
| SBS | Chr2 | 16828699 | T-A | HET | ||
| SBS | Chr2 | 17001269 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g28730 |
| SBS | Chr2 | 4054127 | G-A | Homo | ||
| SBS | Chr2 | 8464518 | G-A | HET | ||
| SBS | Chr3 | 17778537 | G-A | HET | ||
| SBS | Chr3 | 19251318 | A-C | HET | ||
| SBS | Chr3 | 24212446 | G-A | Homo | ||
| SBS | Chr3 | 35576413 | C-G | HET | ||
| SBS | Chr4 | 23596210 | T-C | HET | ||
| SBS | Chr4 | 23917467 | C-T | HET | ||
| SBS | Chr4 | 25933266 | G-A | HET | ||
| SBS | Chr4 | 28223352 | A-G | HET | ||
| SBS | Chr4 | 28893437 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g48460 |
| SBS | Chr4 | 29737760 | G-C | Homo | ||
| SBS | Chr4 | 3120940 | T-G | Homo | ||
| SBS | Chr4 | 3158188 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g06080 |
| SBS | Chr4 | 31680269 | A-T | HET | ||
| SBS | Chr4 | 31680273 | A-G | HET | ||
| SBS | Chr4 | 6505943 | C-T | HET | ||
| SBS | Chr6 | 15595452 | T-A | Homo | ||
| SBS | Chr6 | 15860641 | C-T | HET | ||
| SBS | Chr6 | 29040600 | G-A | HET | ||
| SBS | Chr6 | 3937446 | A-G | HET | ||
| SBS | Chr7 | 11542306 | G-A | HET | ||
| SBS | Chr7 | 13724565 | A-T | HET | ||
| SBS | Chr7 | 148209 | T-C | HET | ||
| SBS | Chr7 | 4543598 | T-A | HET | ||
| SBS | Chr7 | 7800486 | C-G | HET | ||
| SBS | Chr8 | 20444446 | G-A | Homo | ||
| SBS | Chr8 | 2398536 | T-G | Homo | ||
| SBS | Chr8 | 687906 | T-C | HET | ||
| SBS | Chr9 | 11679057 | C-T | HET | ||
| SBS | Chr9 | 1706465 | C-T | HET |
Deletions: 40
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1485983 | 1485984 | 1 | |
| Deletion | Chr5 | 2226967 | 2226969 | 2 | |
| Deletion | Chr7 | 2455168 | 2455171 | 3 | |
| Deletion | Chr7 | 3163956 | 3163959 | 3 | |
| Deletion | Chr12 | 3500564 | 3500575 | 11 | |
| Deletion | Chr5 | 4961001 | 4969000 | 7999 | 2 |
| Deletion | Chr10 | 5336646 | 5336676 | 30 | |
| Deletion | Chr10 | 7763578 | 7763579 | 1 | |
| Deletion | Chr3 | 8082187 | 8082198 | 11 | |
| Deletion | Chr12 | 9082131 | 9082143 | 12 | |
| Deletion | Chr1 | 9099361 | 9099365 | 4 | LOC_Os01g16124 |
| Deletion | Chr1 | 9185076 | 9185078 | 2 | |
| Deletion | Chr2 | 9701403 | 9701404 | 1 | LOC_Os02g16990 |
| Deletion | Chr1 | 10684922 | 10684927 | 5 | |
| Deletion | Chr1 | 10810001 | 10823000 | 12999 | 3 |
| Deletion | Chr4 | 10922750 | 10922751 | 1 | LOC_Os04g19610 |
| Deletion | Chr6 | 11199814 | 11199817 | 3 | |
| Deletion | Chr5 | 12145716 | 12145728 | 12 | |
| Deletion | Chr11 | 13356439 | 13356440 | 1 | LOC_Os11g23200 |
| Deletion | Chr9 | 13720579 | 13720580 | 1 | |
| Deletion | Chr10 | 14389750 | 14389765 | 15 | |
| Deletion | Chr12 | 15008203 | 15008218 | 15 | |
| Deletion | Chr3 | 16325379 | 16325380 | 1 | |
| Deletion | Chr8 | 16399824 | 16399825 | 1 | |
| Deletion | Chr12 | 16434348 | 16434358 | 10 | LOC_Os12g27870 |
| Deletion | Chr5 | 16930125 | 16930127 | 2 | |
| Deletion | Chr11 | 19438677 | 19438678 | 1 | |
| Deletion | Chr2 | 20549654 | 20549669 | 15 | LOC_Os02g34350 |
| Deletion | Chr11 | 20726003 | 20726006 | 3 | |
| Deletion | Chr7 | 21210342 | 21210350 | 8 | |
| Deletion | Chr3 | 21954834 | 21954839 | 5 | |
| Deletion | Chr12 | 22503550 | 22503562 | 12 | |
| Deletion | Chr1 | 22678826 | 22678828 | 2 | |
| Deletion | Chr8 | 22786861 | 22786883 | 22 | |
| Deletion | Chr4 | 24322217 | 24322222 | 5 | |
| Deletion | Chr12 | 26388877 | 26388890 | 13 | |
| Deletion | Chr4 | 32922231 | 32922237 | 6 | |
| Deletion | Chr4 | 33608057 | 33608059 | 2 | |
| Deletion | Chr3 | 33608451 | 33608452 | 1 | LOC_Os03g59040 |
| Deletion | Chr2 | 34924778 | 34924780 | 2 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 20292057 | 20292194 | 138 | |
| Insertion | Chr4 | 9823576 | 9823576 | 1 | LOC_Os04g17880 |
| Insertion | Chr5 | 49552 | 49553 | 2 | |
| Insertion | Chr7 | 634561 | 634565 | 5 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1996919 | 2046844 | 2 |
| Inversion | Chr7 | 1997094 | 2046855 | 2 |
| Inversion | Chr3 | 3744571 | 3744679 | LOC_Os03g07370 |
| Inversion | Chr7 | 21202729 | 21219936 | |
| Inversion | Chr7 | 21203194 | 21220139 | |
| Inversion | Chr5 | 23624714 | 23625131 | LOC_Os05g40220 |
| Inversion | Chr4 | 23635221 | 23635526 | LOC_Os04g39670 |
| Inversion | Chr3 | 32928111 | 32928398 | LOC_Os03g57780 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 13678443 | Chr2 | 16818197 | |
| Translocation | Chr6 | 15405516 | Chr4 | 31987837 | LOC_Os04g53670 |
| Translocation | Chr11 | 27077873 | Chr7 | 21220132 | |
| Translocation | Chr11 | 27077880 | Chr7 | 21219937 |
