Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3715-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3715-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 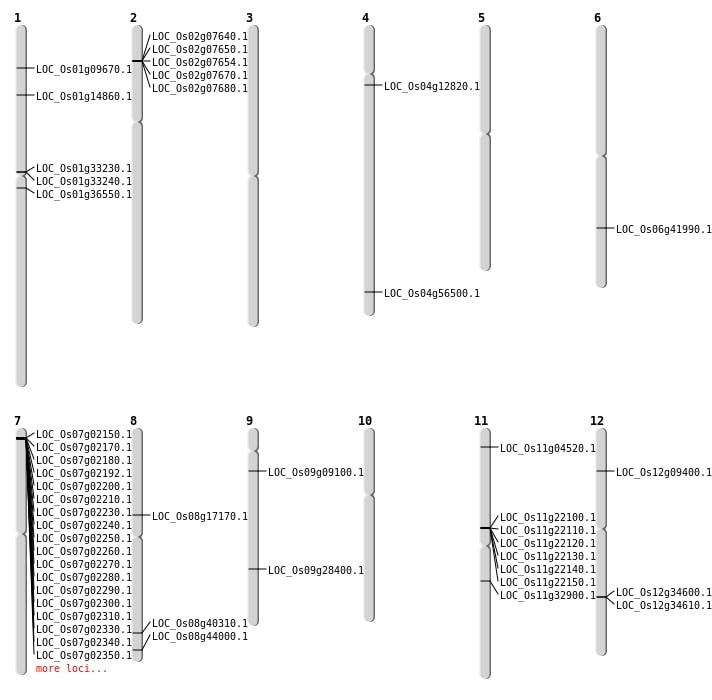
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14932021 | G-A | HET | ||
| SBS | Chr1 | 20151602 | A-G | HET | ||
| SBS | Chr1 | 20287439 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g36550 |
| SBS | Chr1 | 26013917 | T-C | HET | ||
| SBS | Chr1 | 35960229 | G-A | Homo | ||
| SBS | Chr1 | 39564825 | A-T | HET | ||
| SBS | Chr1 | 7463146 | A-G | HET | ||
| SBS | Chr1 | 8333217 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g14860 |
| SBS | Chr10 | 15911545 | C-A | HET | ||
| SBS | Chr10 | 18026260 | C-A | HET | ||
| SBS | Chr10 | 20986732 | C-T | HET | ||
| SBS | Chr10 | 22815472 | T-A | HET | ||
| SBS | Chr10 | 4437110 | C-G | HET | ||
| SBS | Chr11 | 18639097 | G-A | HET | ||
| SBS | Chr11 | 19434257 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g32900 |
| SBS | Chr11 | 5488045 | A-T | HET | ||
| SBS | Chr11 | 8787501 | A-C | HET | ||
| SBS | Chr11 | 9851443 | C-T | Homo | ||
| SBS | Chr12 | 21842070 | T-C | HET | ||
| SBS | Chr12 | 4929817 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g09400 |
| SBS | Chr12 | 7349008 | G-A | HET | ||
| SBS | Chr2 | 14618543 | G-A | HET | ||
| SBS | Chr2 | 22708085 | G-A | HET | ||
| SBS | Chr2 | 26249444 | T-C | HET | ||
| SBS | Chr2 | 27779915 | G-T | HET | ||
| SBS | Chr2 | 29332827 | T-A | HET | ||
| SBS | Chr3 | 10239492 | G-A | HET | ||
| SBS | Chr3 | 19418343 | G-T | Homo | ||
| SBS | Chr3 | 25176740 | A-G | Homo | ||
| SBS | Chr3 | 4184679 | T-A | HET | ||
| SBS | Chr4 | 11339432 | T-A | HET | ||
| SBS | Chr4 | 236537 | A-G | Homo | ||
| SBS | Chr4 | 4271371 | C-A | HET | ||
| SBS | Chr4 | 8605518 | G-A | HET | ||
| SBS | Chr5 | 11533062 | C-T | Homo | ||
| SBS | Chr5 | 89936 | G-A | HET | ||
| SBS | Chr6 | 14331446 | A-G | HET | ||
| SBS | Chr6 | 17639993 | T-C | HET | ||
| SBS | Chr6 | 25210040 | C-A | HET | ||
| SBS | Chr6 | 25210041 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g41990 |
| SBS | Chr6 | 3141570 | C-G | Homo | ||
| SBS | Chr6 | 994255 | G-A | Homo | ||
| SBS | Chr7 | 1602539 | C-T | HET | ||
| SBS | Chr7 | 20211383 | A-T | Homo | ||
| SBS | Chr7 | 8546411 | C-T | Homo | ||
| SBS | Chr8 | 16227174 | G-C | HET | ||
| SBS | Chr8 | 17986257 | G-A | HET | ||
| SBS | Chr8 | 19390030 | T-A | HET | ||
| SBS | Chr8 | 27699635 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g44000 |
| SBS | Chr8 | 8187614 | T-C | HET | ||
| SBS | Chr9 | 17290128 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g28400 |
| SBS | Chr9 | 4867313 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g09100 |
| SBS | Chr9 | 9676857 | G-T | Homo |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 688001 | 874000 | 185999 | 33 |
| Deletion | Chr7 | 885001 | 955000 | 69999 | 13 |
| Deletion | Chr6 | 3768167 | 3768179 | 12 | |
| Deletion | Chr2 | 3959001 | 4002000 | 42999 | 5 |
| Deletion | Chr12 | 4728059 | 4728060 | 1 | |
| Deletion | Chr3 | 5282317 | 5282319 | 2 | |
| Deletion | Chr4 | 7082544 | 7082545 | 1 | LOC_Os04g12820 |
| Deletion | Chr3 | 9889227 | 9889239 | 12 | |
| Deletion | Chr1 | 10370889 | 10370891 | 2 | |
| Deletion | Chr8 | 10521500 | 10521504 | 4 | LOC_Os08g17170 |
| Deletion | Chr11 | 12690001 | 12706000 | 15999 | 6 |
| Deletion | Chr3 | 14884106 | 14884116 | 10 | |
| Deletion | Chr6 | 17499979 | 17499982 | 3 | |
| Deletion | Chr12 | 17923304 | 17923314 | 10 | |
| Deletion | Chr8 | 20575137 | 20575139 | 2 | |
| Deletion | Chr12 | 20969001 | 20983000 | 13999 | 2 |
| Deletion | Chr11 | 23726883 | 23726898 | 15 | |
| Deletion | Chr11 | 24049107 | 24049118 | 11 | |
| Deletion | Chr8 | 25524893 | 25524900 | 7 | LOC_Os08g40310 |
| Deletion | Chr2 | 25648139 | 25648140 | 1 | |
| Deletion | Chr8 | 26190001 | 26190010 | 9 | |
| Deletion | Chr8 | 26831456 | 26831458 | 2 | |
| Deletion | Chr2 | 32334814 | 32334822 | 8 | |
| Deletion | Chr4 | 32705973 | 32705985 | 12 | |
| Deletion | Chr2 | 33527278 | 33527287 | 9 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 21379748 | 21379769 | 22 | |
| Insertion | Chr5 | 29249791 | 29249791 | 1 | |
| Insertion | Chr6 | 23088753 | 23088753 | 1 |
Inversions: 11
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1279555 | 1870094 | |
| Inversion | Chr7 | 1279559 | 1870095 | |
| Inversion | Chr1 | 1895463 | 1895732 | |
| Inversion | Chr1 | 4982251 | 5159122 | LOC_Os01g09670 |
| Inversion | Chr1 | 4982254 | 5159127 | LOC_Os01g09670 |
| Inversion | Chr7 | 10724598 | 10724754 | LOC_Os07g18120 |
| Inversion | Chr1 | 17433860 | 18302220 | LOC_Os01g33230 |
| Inversion | Chr1 | 17433867 | 18306961 | LOC_Os01g33240 |
| Inversion | Chr3 | 21107928 | 21108143 | |
| Inversion | Chr6 | 24491556 | 25148078 | |
| Inversion | Chr6 | 24491571 | 25148127 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 327627 | Chr4 | 11184837 | |
| Translocation | Chr11 | 1904348 | Chr4 | 33677755 | 2 |
| Translocation | Chr11 | 1905779 | Chr1 | 18302219 | 2 |
| Translocation | Chr4 | 33677754 | Chr1 | 18306966 | 2 |
