Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3838-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3838-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 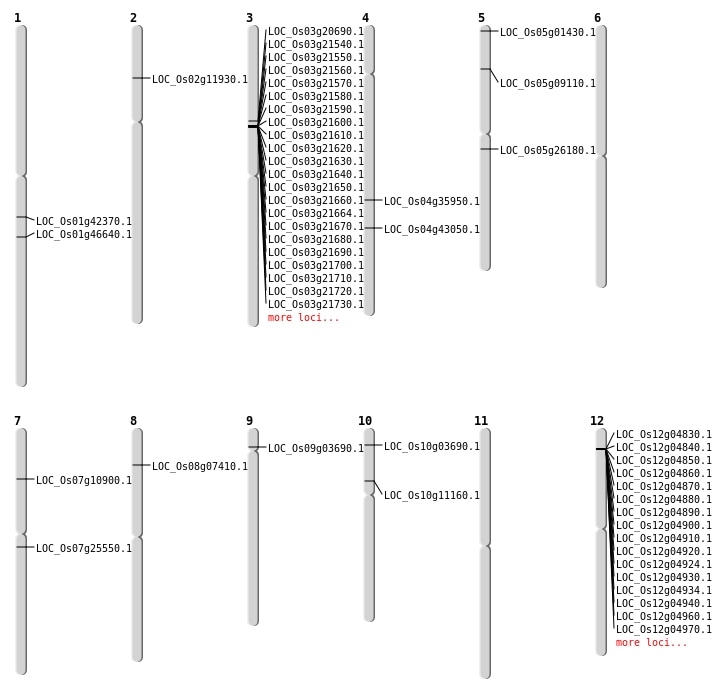
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24072577 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g42370 |
| SBS | Chr1 | 39730126 | C-A | HET | ||
| SBS | Chr1 | 40893093 | C-T | HET | ||
| SBS | Chr10 | 1658670 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g03690 |
| SBS | Chr10 | 6183298 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g11160 |
| SBS | Chr11 | 15175928 | G-A | HET | ||
| SBS | Chr11 | 24057797 | C-G | Homo | ||
| SBS | Chr11 | 3197898 | G-T | HET | ||
| SBS | Chr12 | 344706 | G-C | HET | ||
| SBS | Chr12 | 4621959 | T-C | Homo | ||
| SBS | Chr2 | 12678767 | T-A | HET | ||
| SBS | Chr2 | 12678777 | T-C | HET | ||
| SBS | Chr2 | 17259792 | C-A | Homo | ||
| SBS | Chr2 | 34186144 | G-A | HET | ||
| SBS | Chr2 | 4798057 | A-G | HET | ||
| SBS | Chr2 | 8208532 | C-A | HET | ||
| SBS | Chr3 | 10131412 | C-G | HET | ||
| SBS | Chr3 | 11706481 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g20690 |
| SBS | Chr3 | 19100921 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33420 |
| SBS | Chr3 | 20164922 | T-A | HET | ||
| SBS | Chr3 | 2572393 | C-A | HET | ||
| SBS | Chr4 | 21920965 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g35950 |
| SBS | Chr4 | 25481163 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43050 |
| SBS | Chr4 | 32825756 | G-A | HET | ||
| SBS | Chr5 | 12665933 | T-A | HET | ||
| SBS | Chr5 | 14006472 | T-A | HET | ||
| SBS | Chr5 | 15237124 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g26180 |
| SBS | Chr5 | 18916693 | C-A | HET | ||
| SBS | Chr5 | 24869796 | T-G | HET | ||
| SBS | Chr5 | 29659755 | T-C | HET | ||
| SBS | Chr5 | 5074213 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09110 |
| SBS | Chr6 | 31222648 | C-T | Homo | ||
| SBS | Chr6 | 6332191 | G-C | HET | ||
| SBS | Chr6 | 8763746 | A-G | HET | ||
| SBS | Chr7 | 1437986 | G-C | Homo | ||
| SBS | Chr7 | 14645129 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g25550 |
| SBS | Chr7 | 19050411 | C-A | HET | ||
| SBS | Chr7 | 22725969 | G-A | Homo | ||
| SBS | Chr7 | 5963369 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g10900 |
| SBS | Chr7 | 9801555 | C-T | HET | ||
| SBS | Chr8 | 14024680 | G-A | HET | ||
| SBS | Chr8 | 14368835 | T-A | HET | ||
| SBS | Chr8 | 23233978 | G-A | Homo | ||
| SBS | Chr8 | 27330659 | G-C | HET | ||
| SBS | Chr8 | 4162163 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g07410 |
| SBS | Chr8 | 5763547 | C-A | HET | ||
| SBS | Chr9 | 13492133 | C-A | HET | ||
| SBS | Chr9 | 16194153 | T-A | HET | ||
| SBS | Chr9 | 1862146 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g03690 |
| SBS | Chr9 | 19929999 | G-C | HET | ||
| SBS | Chr9 | 4645796 | C-T | Homo | ||
| SBS | Chr9 | 4645797 | C-G | Homo | ||
| SBS | Chr9 | 5682052 | T-C | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 248502 | 248507 | 5 | LOC_Os05g01430 |
| Deletion | Chr12 | 2071001 | 2495000 | 423999 | 62 |
| Deletion | Chr12 | 2497001 | 2587000 | 89999 | 17 |
| Deletion | Chr12 | 3483693 | 3483694 | 1 | |
| Deletion | Chr4 | 3942825 | 3942831 | 6 | |
| Deletion | Chr11 | 3976042 | 3976043 | 1 | |
| Deletion | Chr7 | 7241624 | 7241626 | 2 | |
| Deletion | Chr6 | 11613757 | 11613759 | 2 | |
| Deletion | Chr3 | 12311001 | 12487000 | 175999 | 32 |
| Deletion | Chr3 | 14943061 | 14943066 | 5 | |
| Deletion | Chr1 | 16780372 | 16780373 | 1 | |
| Deletion | Chr1 | 16945452 | 16946101 | 649 | |
| Deletion | Chr7 | 18914708 | 18914710 | 2 | |
| Deletion | Chr8 | 21922302 | 21922305 | 3 | |
| Deletion | Chr6 | 23194508 | 23194509 | 1 | |
| Deletion | Chr8 | 23681849 | 23681857 | 8 | |
| Deletion | Chr1 | 26560084 | 26560091 | 7 | LOC_Os01g46640 |
| Deletion | Chr1 | 27197998 | 27198003 | 5 | |
| Deletion | Chr3 | 27639324 | 27639331 | 7 | |
| Deletion | Chr6 | 29710279 | 29710280 | 1 | |
| Deletion | Chr1 | 31761061 | 31761067 | 6 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 14908542 | 14908723 | 182 | |
| Insertion | Chr4 | 22948783 | 22948785 | 3 | |
| Insertion | Chr9 | 331362 | 331367 | 6 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 6185173 | 6221496 | LOC_Os02g11930 |
| Inversion | Chr2 | 6185178 | 6221530 | LOC_Os02g11930 |
| Inversion | Chr6 | 22483108 | 22521710 | |
| Inversion | Chr8 | 27301247 | 27301429 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2147144 | Chr11 | 5073923 | |
| Translocation | Chr6 | 18190425 | Chr3 | 8985228 |
